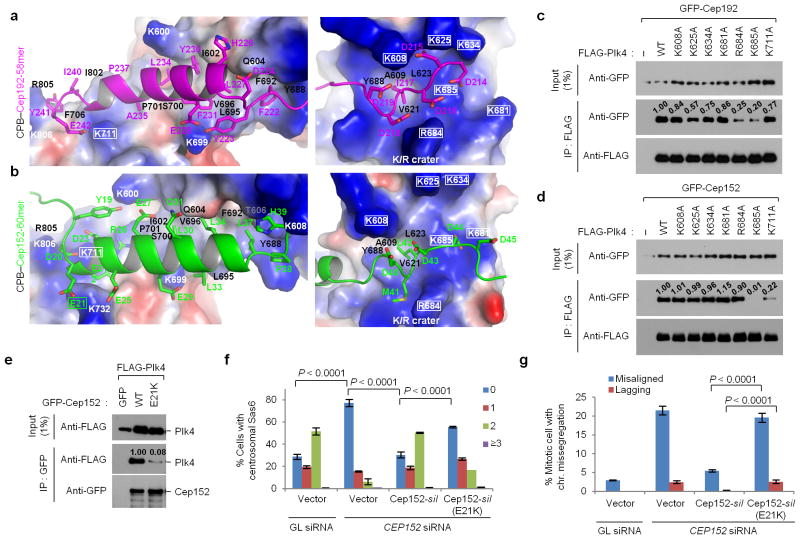
Figure 3.
The binding modes of Cep192-58mer and Cep152-60mer to Plk4 CPB and mutational analyses of residues critical for the interaction. (a,b) Electrostatic surface representation of the CPB–58mer (a) and the CPB –60mer (b) complexes. The K711 and K/R crater residues of CPB and the E21 of Cep152-60mer are highlighted in rectangles. See Supplementary Figure 3e – h for details. (c–e) Immunoprecipitation (IP) analyses of 293T cells cotransfected with the indicated constructs. –, control vector; numbers, relative signal intensities. (f, g) Characterization of a cancer-associated CEP152 E21K mutation. U2OS cells stably expressing either control vector or siRNA-resistant CEP152-silor CEP152-silE21K mutant were silenced for either control Luciferase (GL siRNA) or CEP152 (CEP152 siRNA) (see Supplementary Table 2), and then immunostained (see Supplementary Figure 4a–c). The numbers of centrosomal Sas6 signals (from 0 to ≥3) among interphase cells (f) and the percentage of mitotic cells with missegregating chromosomes (chr.) (g) were quantified from three independent experiments (≥200 cells/cell line/experiment). Error bars, s.d. Uncropped blot images for c–e are shown in Supplementary Figure 8c–e.