Fig. 2.
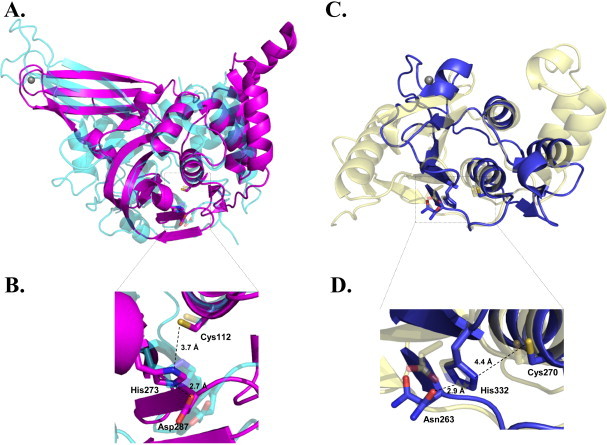
Structures of SARS-CoV PLpro and EAV PLP2 compared to USP14 and yeast OTU1. (A) Structural superposition of SARS-CoV PLpro (magenta, PDB code 2FE8) to USP14 (cyan, PDB code 2AYO). The zinc atom from the zinc finger of SARS-CoV PLpro is shown as a gray sphere. (B) Close-up view of the active site in SARS-CoV PLpro with the catalytic triad residues shown as sticks. Numbering of the residues is based on (26) (C) Structural overlay of EAV PLP2 (blue, PDB code 4IUM) with yeast OTU1 (yellow, PDB code 3BY4) bound to ubiquitin. The ubiquitin molecules have been omitted for clarity. The zinc atom from the zinc finger of EAV PLP2 is shown in gray sphere. (D) Close-up view of the active site in EAV PLP2. Numbering of the residues is based on (49). Asn263 in EAV PLP2 catalytic triad has two alternative conformations. Images were generated using PyMOL.
