Fig. 3.
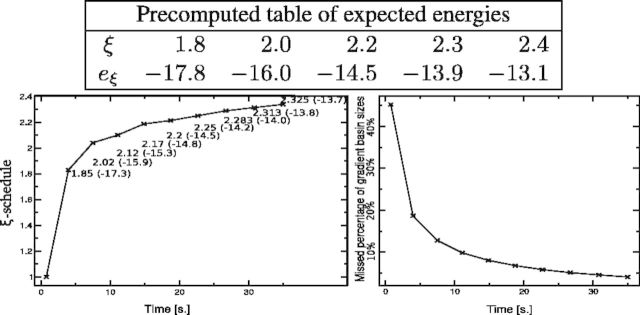
Computation of LMs for Melitaea cinxia U6 snRNA JX878560.1 (107 nt) with RNAlocmin. (Left) Adaptive ξ schedule as a function of run time. For each sampling epoch we show the values of e and ξ as 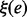 . The precomputed
. The precomputed 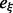 table is shown at the top. (Right) Size-weighted fraction of undiscovered LMs compared with exhaustive enumeration with RNAsubopt and barriers. Basin sizes are estimated from the 106 structures with the lowest energies
table is shown at the top. (Right) Size-weighted fraction of undiscovered LMs compared with exhaustive enumeration with RNAsubopt and barriers. Basin sizes are estimated from the 106 structures with the lowest energies
