Table 1.
Performance comparison with different folding path prediction algorithms for the refolding paths between the MFE structure and a randomly selected LM
| Algorithm | Number of best runs |
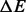 (kcal/mol) (kcal/mol) |
Time (s) |
|---|---|---|---|
| RNAtabupath | 14 | 3.0598 | 4617.7 |
| BHGbuild | 34 | 1.1028 | 7.6674 |
| BHGbuild -noF | 34 | 1.1028 | 0.6824 |
| Pathfinder | 95 | 0.0367 | 113.01 |
| findpath | 12 | 1.5104 | 0.6397 |
Note: Values are averages over 100 RNA sequences of length 200 nt. 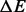 is the average difference in the energy from the best run. BHGbuild -noF is the BHG algorithm without the optional flooding step. Pathfinder was run with option -M DB-MFE, and for findpath we used depth
=
1000.
is the average difference in the energy from the best run. BHGbuild -noF is the BHG algorithm without the optional flooding step. Pathfinder was run with option -M DB-MFE, and for findpath we used depth
=
1000.
