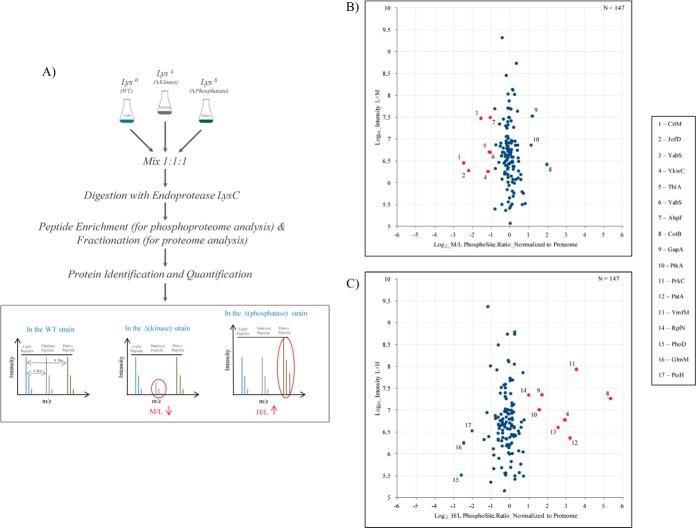
Fig. 4.
SILAC screen of potential PrkC and PrpC substrates. A, Overview of the proteomics workflow adopted for the identification of novel substrates for the Ser/Thr kinase PrkC and Ser/Thr phosphatase PrpC. Briefly, cells were harvested at mid stationary phase. Extracted proteins were digested and fractionated by Offgel or GeLC. Portion of the digested peptides was analyzed for phosphorylation sites as before. WT acts as a control and all ratios were normalized to this state. Proteins with a down regulated M/L ratio and/or an up-regulated H/L ratio were considered as potential candidates for PrkC and PrpC respectively. B, C, Scatter plot showing; ΔprkC/WT SILAC ratios and ΔprpC/WT SILAC ratios respectively. Log2 ratios of the phosphorylation sites are normalized to the corresponding protein and plotted against peptide intensities in Log10 scale. Significantly changing (p = 0.05) SILAC ratios marked in red, present potential substrates of PrkC or PrpC respectively and those marked in green are differentially regulated but not direct substrates of these enzymes.