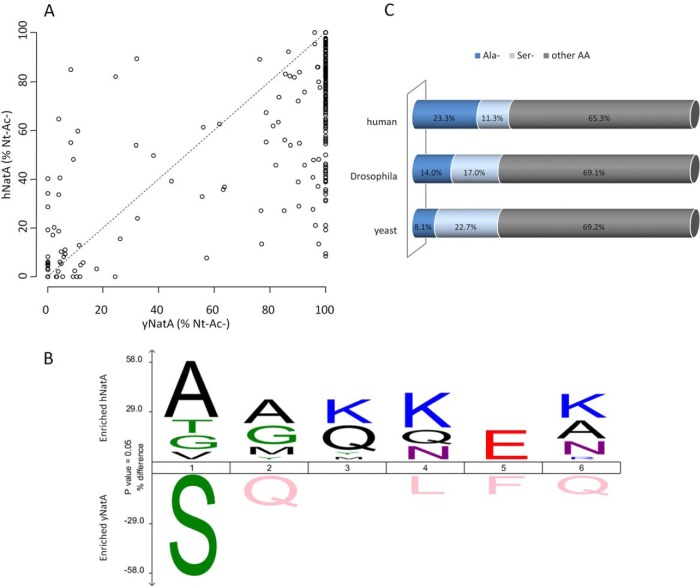
Fig. 5.
Species-specific NatA substrate specificity profiles indicate a co-evolution of NatA complexes with their species-matching substrate pools. A, Scatterplot displaying the correlation of the degrees of Nt-acetylation of the NatA type class N termini (i.e. Ser-, Ala-, Thr-, Val-, and Gly- starting N termini). Correlation of the degrees of Nt-acetylation of the determined NatA type class N termini identified in the yeast N-terminomes of a yeast NatA (yNatA) (x-axis) and a human NatA (hNatA) expressing yeast strain (y-axis) (n = 333). In 11.7% of the cases, hNatA expression results in an increase of Nt-Ac as compared with the yNatA whereas in 27.6% of the cases about equal levels of Nt-Ac (less than 10% difference in the degree of Nt-Ac) could be observed. For 60.7% of identified NatA-substrates, hNatA expression resulted in a lower level of Nt-Ac as compared with the levels in control yeast (yNatA). B, A differential iceLogo (63) representation was created using two y/hNatA substrate subsets for which the hNatA (n = 30) or yNatA (n = 30) Nt-acetylation efficiency differed most notably (as deduced from the proteomics data and based on the degrees of Nt-acetylation calculated), thus representing the enriched and depleted residues in these respective NatA substrate sets. Statistically significant residues (p ≤ 0.05) are plotted with the size of the amino acid proportional to the difference observed in Nt-acetylation efficiency between human and yeast NatA. Multiple sequence alignments are given as 1 to 6 corresponding to the first 6 N-terminal residues of NatA substrates and with Nt-acetylation occurring at position 1. C, Stacked horizontal bar charts of the differential alanine and serine occurrence at position 2 of yeast (S. cerevisiae), fruit fly (D. melanogaster), and human (H. sapiens) iMet starting Swiss-Prot database protein entries (respectively 6620, 3168 and 20114 reviewed entries in the UniProtKB/Swiss-Prot protein database release 2013_07).