Figure 1. Two euAGAMOUS genes in Medicago truncatula.
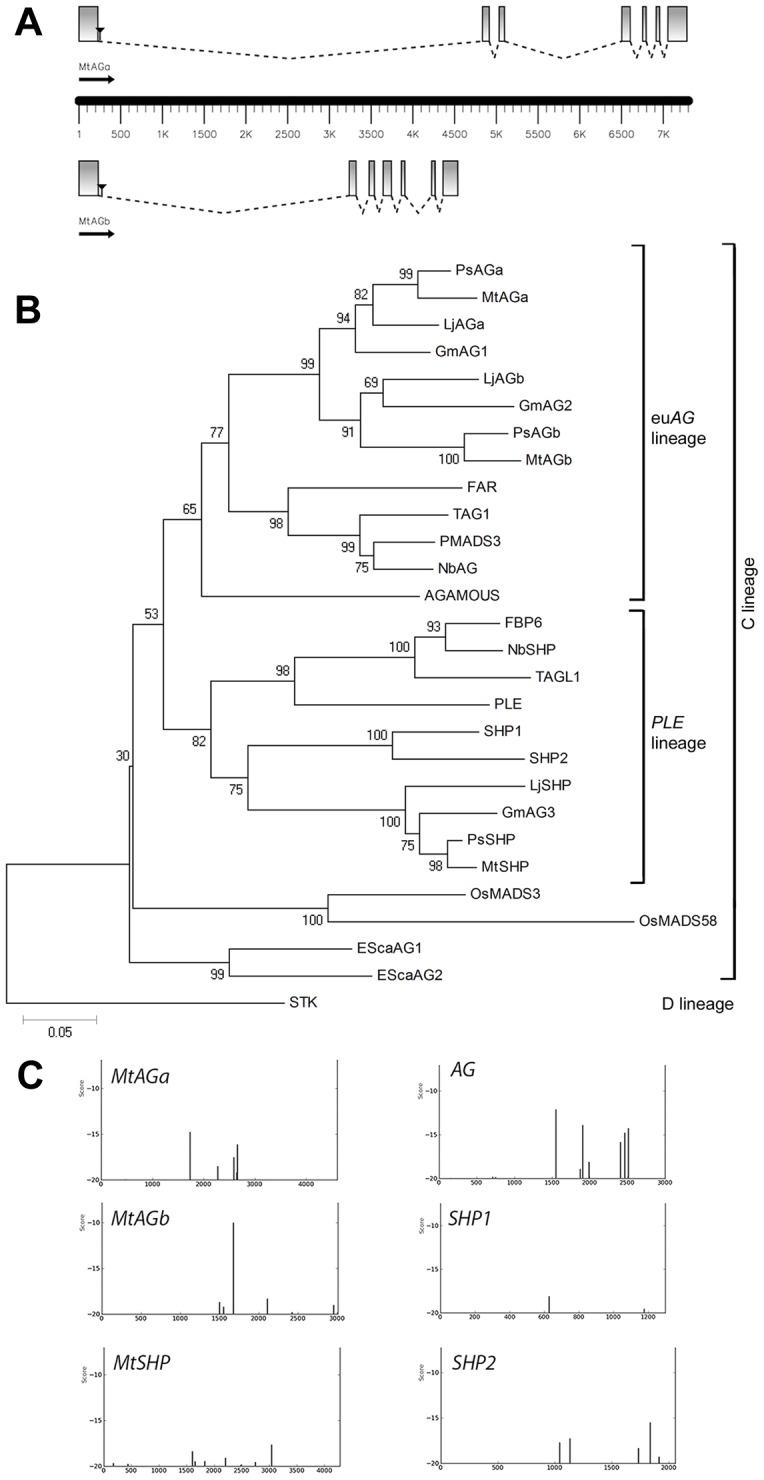
(A) Gene structure of MtAGa and MtAGb. Coding sequences are represented as boxes and introns as dotted lines. Black triangles localize the position of the Tnt1 insertions present in the mutant lines mtaga (NF13380) and mtagb (NF4908) used in this study. (B) Neighbor-Joining Tree of euAG and PLENA homologs from a selection of diverse species. The numbers next to the nodes refer to bootstrap values from 10000 pseudo-replicates. (C) Distribution of putative LFY binding sites in the first intron of MtAGa, MtAGb and MtSHP genes as identified with the use of a position-specific scoring matrix using a cutoff value of −20.
