Figure 2. Sdh6 or Sdh7 functions are linked to the Fe/S Sdh2 subunit.
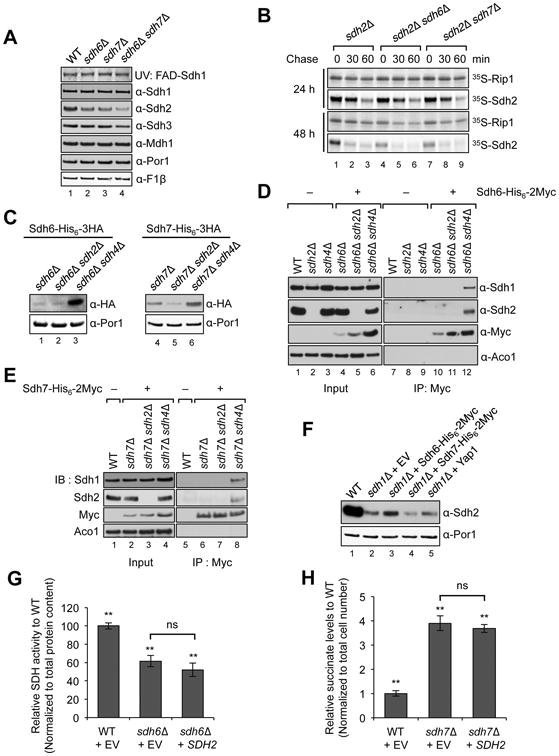
(A) FAD-containing Sdh1 was visualized by UV excitation. Other proteins visualized by immunoblotting include Mdh1, malate dehydrogenase and Por1, porin (loading control). (B) 35S-methionine labeled proteins were incubated with isolated mid-log vs. stationary phase mitochondria for 30 min (pulse), followed by blocking protein import with valinomycin for 30 and 60 min (chase), respectively. Radiolabeled proteins were resolved on SDS-PAGE and detected by autoradiography. (C) Sdh6-His6-3HA or Sdh7-His6-3HA under their own endogenous promoters was expressed from plasmids in cells lacking either Sdh2 or Sdh4 along with endogenous Sdh6 or Sdh7 depleted, respectively. Steady-state levels are shown by immunoblotting. (D) Coimmunoprecipitation of Sdh6-His6-2Myc after crosslinking. Mitochondria were solubilized with 1% digitonin in the presence of 1 mM dithiobis[succinimidylpropionate]. The cross-linking reaction was stopped with Tris buffer (pH 7.4), and the supernatants were absorbed to anti-Myc antibody-conjugated magnetic beads. Bound substances to Myc beads were resolved on SDS-PAGE and detected by immunoblotting. Input, 4% of total lysates; Aco1, FeS aconitase. (E) Standard co-immunoprecipitation of Sdh7-His6-2Myc was performed with isolated mitochondria without crosslinking. Input, 2% of total lysates. (F) Steady-state levels of Sdh2 in sdh1Δ mutants with overexpression of proteins indicated. Yap1, transcription factor up-regulating oxidative stress response genes. (G) SDH activity in sdh6Δ mutants with SDH2 overexpression was detected as described in Figure 1E. Data are represented as mean ± SD (N=3; **p < 0.05; ns, not significant). (H) Succinate levels in sdh7Δ mutants with SDH2 overexpression were measured as described in Figure 1D. Mean ± SEM is shown (N=6; **p < 0.05; ns, not significant). See also Figure S3.
