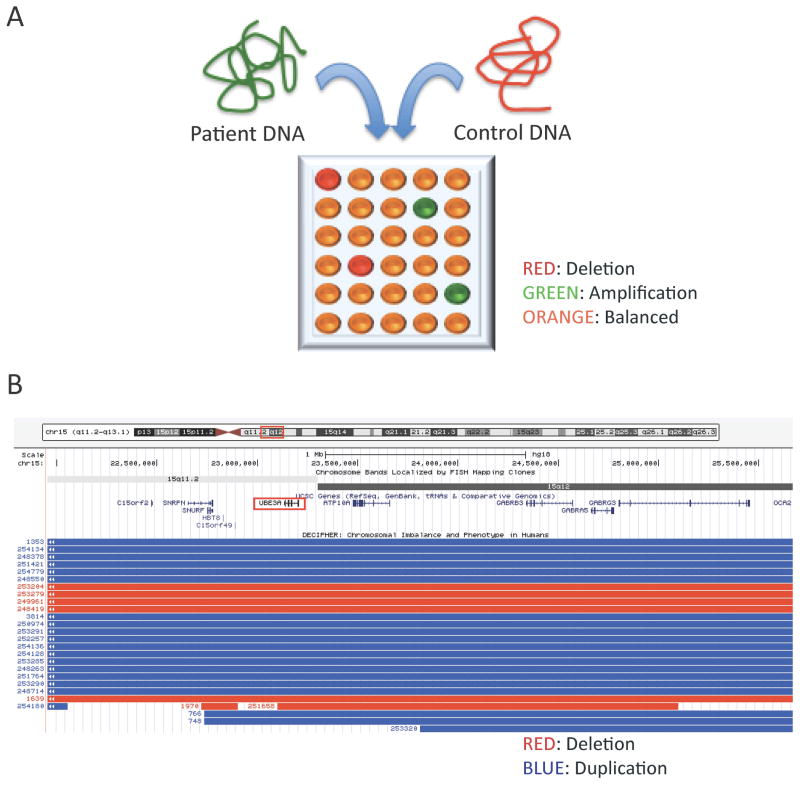
Figure 1. Comparative Genomic Hybridization (CGH) assays frequently reveal copy number variants (CNVs) in patients with unexplained epileptic encephalopathy.
(A) Comparative Genomic Hybridization (CGH) assays are conducted by comparing a patient’s genomic DNA, labeled with a fluorescent dye such as fluorescein (green), to a control DNA, labeled with another fluorescent dye such as rhodamine (red), both applied on a microarray chip in which each well contains a probe specific for a given genomic interval. Deletions (red) and amplifications (green) in the proband’s DNA can be differentiated from areas with balanced (normal) DNA. The sensitivity of CGH analysis is inversely proportional to the spacing of consecutive probes, usually around 500Kb. (B) Examples of microdeletions (red) and microduplications (blue) presenting with neurodevelopmental phenotypes and reported in the Angelman’s syndrome interval 15q11-13, encompassing the UBE3A gene (generated using the UCSC Genome Browser on March 17th, 2014).