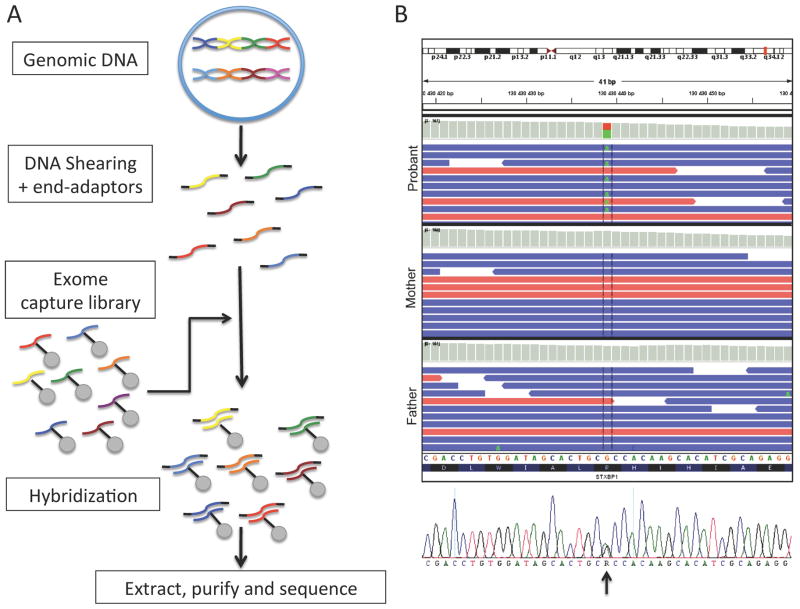
Figure 2. Whole exome sequencing often reveals detrimental de novo mutations in patients with sporadic epileptic encephalopathies.
(A) Schematic representation of the procedure involved in conducting whole exome sequencing. The proband’s genomic DNA is first sheared in ≈200bp fragments which are protected with end-adaptors. Secondly, the patient’s DNA is hybridized to an exome capture library consisting of specific probes designed to recognize most coding fragments of human DNA (i.e. exons and adjacent intron-exon splice-site junctions). The hybridized fragments are extracted using systems such as streptavidin-labeled beads. The patient’s exonic DNA is then retrieved and sent for sequencing (massive parallel sequencing). (B) The sequences obtained are aligned to the reference human genome sequence. Multiple reads will be obtained for each genomic interval sequenced. De novo variants that are unique to the proband and not inherited from the parents can be identified. These variants are then confirmed using Sanger re-sequencing (illustrated in bottom panel). In this particular patient presenting with early-onset epileptic encephalopathy, exome sequencing revealed a single de novo variant, where an A replaces the reference G in a heterozygous fashion (c. G875A), in the well-known epileptic encephalopathy gene STXBP1. This variant was predicted pathogenic by different bioinformatic scores, such as SIFT and PolyPHEN, which consider the variant’s impact on protein structure and domain conservation.