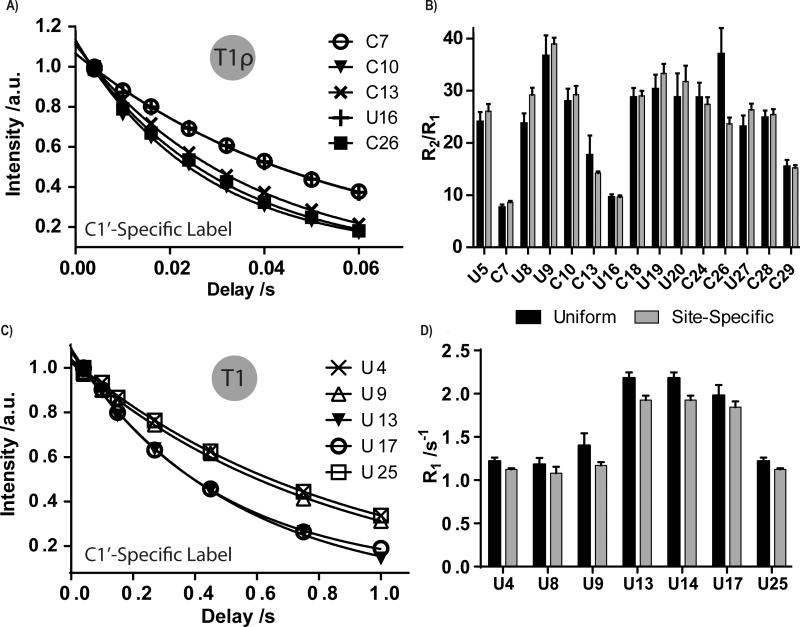
Figure 4.
Site-specific labeling improves dynamics measurements of the IRE and ESS3 RNAs. (a,c) Curve fits of T1ρ or T1 experiments on five residues of the IRE RNA and ESS3 RNA, respectively. (b,d) Comparison of overall dynamics (R2/R1 ratio or R1 values) of all U and/or C residues of IRE and ESS3 RNAs, respectively, when the molecule was 13C/15N uniformly or site-specifically labeled. In the IRE RNA, U8, C13, and C26 show significant differences in R2/R1 ratios. In ESS3 RNA, U9, U13, and U14 show significant differences in R1 decay rates. Other residues remain unaffected. Errors bars are shown as standard deviation from three independent experiments.