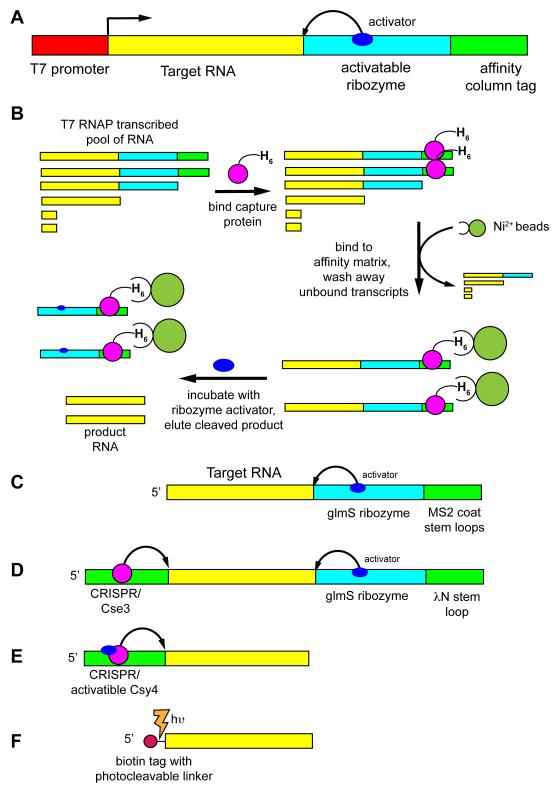
Figure 2.
Schematic of various native affinity tag purification methods. (A) The general scheme involves creating a dsDNA template for use in a T7 RNAP transcription reaction that contains a promoter element (red), the RNA of interest on the 5′-end of the transcript (yellow), an activatable ribozyme that cleaves at the 3′-end of the RNA of interest (cyan) and an affinity tag at the extreme 3′-end of the transcript (green) for selective immobilization of the full-length transcript to a column. (B) Purification of the RNA of interest in the variation using a hexahistidine-tagged MS2 coat binding protein comprises adding the capture protein to the transcription reaction and allowing it to bind, passing the reaction over a nickel affinity column to bind full-length transcript, washing away other transcription reaction components, followed by eluting the RNA of interest by incubating the column with an effector that activates the ribozyme [22,37]. Variations of this methodology use different combination of cleavage methods and tags, including (C) glmS/MS2 coat [37], (D) the “ARiBo” tag using both a 3′- glmS/λN peptide module and a 5′ Cse3 tag [38], (E) an activatable Cys4-binding module at the 5′-end that is used for both immobilization and cleavage [36], and (F) a 5′ photocleavable biotin tag [46].