Fig. 6.
Mechanisms for division apparatus localization. a
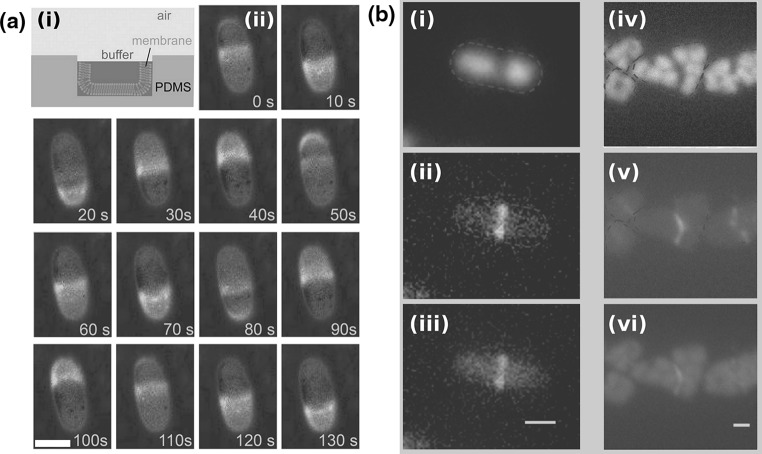
i Assay for the reconstitution of MinDE in microchambers (side view). Microchambers are fabricated in PDMS, coated with a supported lipid bilayer, and buffer with MinD, MinE and ATP is placed on top. ii
Top-view images of MinDE dynamic localization in one of the microchambers of a. Time is in seconds; 10 % of the MinE proteins are labeled with Alexa-488. Scale bar 10  . b Localization of the nucleoid and FtsZ-GFP in rod shaped (i–iii
left column) and anomalously shaped E. coli cells (iv–vi
right column). Nucleoid is labeled with the nucleoid-associated protein HupA conjugated to RFP. Red dashed line represents the outline of the cells. i, iv Chromosomes profile. ii, v FtsZ-GFP. iii, vi Overlay of the HupA-RPF (orange) and the FtsZ-GFP (green) images. Scale bar in both cases is 1
. b Localization of the nucleoid and FtsZ-GFP in rod shaped (i–iii
left column) and anomalously shaped E. coli cells (iv–vi
right column). Nucleoid is labeled with the nucleoid-associated protein HupA conjugated to RFP. Red dashed line represents the outline of the cells. i, iv Chromosomes profile. ii, v FtsZ-GFP. iii, vi Overlay of the HupA-RPF (orange) and the FtsZ-GFP (green) images. Scale bar in both cases is 1  . a Modified with permission from reference Zieske and Schwille (2013)
. a Modified with permission from reference Zieske and Schwille (2013)  (2013) John Wiley and Sons, Inc. b Modified with permission from reference Männik et al. (2012)
(2013) John Wiley and Sons, Inc. b Modified with permission from reference Männik et al. (2012)  (2012) National Academy of Sciences, USA. (Color figure online)
(2012) National Academy of Sciences, USA. (Color figure online)