Abstract
The risk of developing hormone-dependent cancers with long-term exposure to estrogens is attributed both to proliferative, hormonal actions at the estrogen receptor (ER), and chemical carcinogenesis elicited by genotoxic, oxidative estrogen metabolites. Non-tumorigenic MCF-10A human breast epithelial cells are classified as ER(−) and undergo estrogen-induced malignant transformation. Selective estrogen receptor modulators (SERMs), in use for breast cancer chemoprevention and for post-menopausal osteoporosis, were observed to inhibit malignant transformation, as measured by anchorage-independent colony growth. This chemopreventive activity was observed to correlate with reduced levels of oxidative estrogen metabolites, cellular ROS, and DNA oxidation. The ability of raloxifene, desmethylarzoxifene (DMA), and bazedoxifene to inhibit this chemical carcinogenesis pathway was not shared by 4-hydroxytamoxifen. Regulation of Phase 2 rather than Phase 1 metabolic enzymes was implicated mechanistically: raloxifene and DMA were observed to upregulate sulfotransferase (SULT 1E1) and glucuronidase (UGT 1A1). The results support upregulation of Phase 2 metabolism in detoxification of catechol estrogen metabolites leading to attenuated ROS formation as a mechanism for inhibition of malignant transformation by a subset of clinically important SERMs.
Keywords: Estrogen metabolism, SERMs, oxidative stress, malignant transformation, breast cancer chemoprevention
Introduction
Breast cancer is the leading cause of cancer death among women in western countries. The association of hormone dependent cancer with exposure to endogenous estrogens has been known for decades. Of the two major mechanisms of estrogen carcinogenesis, the hormonal pathway, mediated via estrogen receptors (ER), has been extensively studied (1-4). Formation of highly reactive estrogen quinone metabolites, which can cause DNA damage, is believed to be a major contributor to chemical carcinogenesis (5-7).
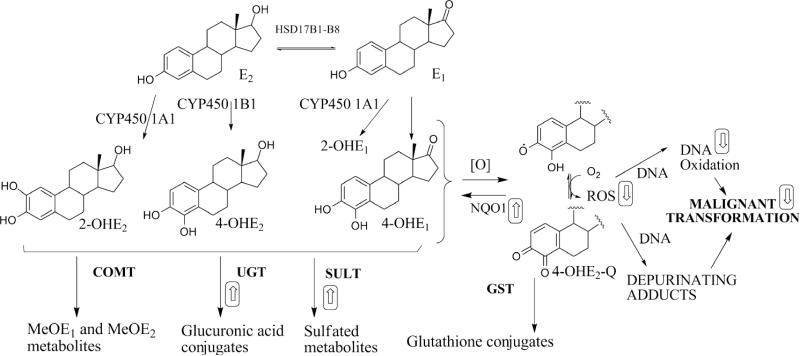
In breast epithelial cells, the endogenous estrogens are metabolized to their 2-OH and 4-OH catechol metabolites, catalyzed by CYP450 1A1 and CYP450 1B1, respectively (Figure 1). Further oxidation of estrogen catechols to quinones causes genotoxicity through electrophilic and oxidative DNA damage, including formation of 8-oxo-7,8-dehydro-2’-deoxyguanosine (8-oxo-dG) (8-10). Formation of reactive oxygen species (ROS) from quinone redox cycling can amplify DNA damage (11, 12). Several lines of evidence strongly suggest that the estrogen catechols are the proximal carcinogens in chemical carcinogenesis (13-17). Prevention of estrogen-induced chemical carcinogenesis therefore can theoretically be achieved by “detoxification” of estrogen catechols, via: 1) attenuated formation; 2) enhanced conjugative metabolism and clearance; or 3) trapping of quinones and ROS (18) (Figure 1) .
Figure 1.
Estrogen metabolism and its relationship to chemical carcinogenesis. In breast epithelial cells, several SERMs were observed to modulate estrogen metabolism (as depicted by boxed arrows), in particular via modulation of detoxification enzymes: sulfotransferase, SULT; UDP-glucuronosyltransferase, UGT; NAPDH:quinone oxidoreductase, NQO1; glutathione-S-transferase-P1, GST. Catechol-O-methyl transferase (COMT) activity was not perturbed by SERMs. SERMs did not modulate expression of CYP450s in E2-treated cells.
Model systems for study of chemical carcinogenesis, a process envisioned to develop over many years of exposure to genotoxic insult, represent a challenge. MCF-10 cells are nontumorigenic human breast epithelial cells that undergo estrogen-induced malignant transformation. Owing to low ER levels and lack of proliferative response to estrogens, the cell line is of use in studying chemical carcinogenesis, in the absence of confounding hormonal proliferative signals (17, 19, 20).
Selective estrogen receptor modulators (SERMs) are ER ligands that oppose the effects of endogenous estrogens in breast tissues . In the present study, the potential for prevention of estrogen-induced malignant transformation of MCF-10A cells was studied in response to raloxifene (Ral), and related SERMs. Ral and DMA, the active metabolite of arzoxifene, were observed to inhibit malignant transformation.
The interconversion of estradiol (E2) with estrone (E1) is catalyzed by the enzyme 17β– hydroxysteroid dehydrogenase (17β-HSD) (Figure 1). In MCF-10A cells, since i) the equilibrium lies strongly towards E1 and ii) the stability of the methyl ether metabolites is superior to the catechol estrogen itself, MeOE1 represents a reliable, indirect measurement of estrogen oxidative metabolism (21). For three SERMs, inhibition of malignant transformation of MCF10-A cells was observed to correlate with attenuation of estrogen metabolism as measured by MeOE1. In order to explain these observations, the response to SERMs of mediators of estrogen Phase 1 and Phase 2 metabolism was studied. “Detoxification” of the catechol estrogen may be mediated by conjugative metabolism by sulfotransferase (SULT), UDP-glucuronosyltransferase (UGT), catechol-O-methyl transferase (COMT), and glutathione-S-transferase (GST), or arguably by NAD(P)H:quinone oxidoreductase (NQO1) (22, 23). Although the expression of UGT is prominent in hepatic tissues (24), in extra-hepatic tissues such as breast, SULT plays a prominent role in detoxification (25, 26). The results indicate that prevention of estrogen-induced transformation by SERMs, resulting from attenuated estrogen metabolism ,is mediated by upregulation of SULT 1E1 and UGT 1A1). Interestingly, of the two further clinical SERMs, bazedoxifene (Baze) and tamoxifen (Tam), Baze attenuated formation of MeOE1, whereas Tam did not. The mechanism of action of Ral and DMA in this model of estrogen-dependent malignant transformation was detoxification of genotoxic estrogen metabolites by upregulation of conjugative metabolism and attenuation of oxidative stress. These observations on non-canonical SERM actions, and the outlier nature of Tam, are of therapeutic relevance for an important drug class.
Materials and Methods
Chemicals and reagents
All chemicals, reagents, and enzymes were obtained from Sigma (St. Louis, MO) or Invitrogen (Carlsbad, CA) unless stated otherwise. Antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA), Cell Signaling Technology (Boston, MA) and Sigma (St. Louis, MO). Chemical standards of estrogen metabolites were obtained from Steraloids Inc. (Newport, RI). 4-Hydroxyestrone-1,2,16,16-d4, and 2-methoxyestrone-1,4,16,16-d4 were obtained from CDN Isotopes (Pointe-Claire, Quebec) and used as internal standards in estrogen metabolism experiments.
Cell lines and cell culture conditions
MCF-10A cells were obtained from American Type Culture Collection (Manassas, VA) and maintained in phenol red and estrogen-free Dulbecco's modified Eagle's medium and F12 medium (DMEM/F12) supplemented with 1% penicillin-streptomycin, 5% fetal bovine serum, cholera toxin (0.1 μg/mL), epidermal growth factor (20 ng/mL), hydrocortisone (0.5 μg/mL), insulin (10 μg/L) and 5% CO2 at 37 °C as described previously (27). MCF-10A cells were authenticated using single tandem repeat analysis (STR). MCF-7 cells were obtained from ATCC and used for standard ERE-luciferase assay as described previously (28). The MDAMB-231:β41 cell line, ER(−) cells stably transfected with ERβ, were a kind gift of Dr D. Tonetti (UIC, Chicago) and used as described for ERE-luciferase assay (29).
Analysis of estrogen metabolites in MCF-10A cells
Estrogen metabolites were analyzed in MCF-10A cell as previously described (27). Briefly, MCF-10A cells were incubated with E2 (1 μM) in the presence or absence of SERMs (1 μM) for 6 days. Treatments were renewed every 3 days. Since, DMA and Ral showed a significant inhibition of estrogen metabolism at 1 μM in MCF-10A cells, a dose response was performed for these two SERMs: DMA and Ral (0.1 - 2.5 μM) were tested in the presence of E2 (1 μM). Sample preparation and analysis was done using the method described by Xu et al. (30) with minor modifications as previously described (27). Enzymatic hydrolysis of cell media was done as previously described (30) with minor modifications. Briefly, MCF-10A cells were plated in 6 well plates with 3 mL of media in each well. Cells were treated with E2 (1 μM) in the presence or absence of SERMs (1 μM) for 6 days and treatments were renewed every 3 days. Cell media was collected every 3 days pooled together to get total of 6 mL for each sample. Standard curves were prepared for 2-MeOE1 and 4-MeOE1 using as internal standard: 2-MeOE1-d4. The internal standard was also added to each sample before further processing. Enzyme hydrolysis buffer was prepared as previously described (30) which contained L-ascorbic acid, β-glucuronidase, and sufatase in 0.15 M sodium acetate buffer (pH= 4.6). Equal amounts (6 mL) of hydrolysis buffer was added into each cell media sample (6 mL) and incubated overnight (16 h) at 37 °C. Samples were extracted into dichloromethane and analyzed using LC/MS-MS as previously described (27). Exemplar amounts of 2-MeOE1 and 4-MeOE1 are provided from standard curves for experiments in which cells were treated with E2 alone: 376 ± 22 pM and 319 ± 95 nM, for 2-MeOE1 and 4-MeOE1, respectively.
ROS formation determined by CM-H2DCFDA
MCF-10A cells were grown (4 × 103 cells/mL) on each of eight chambers on a sterile NuncTM chambered coverglass and incubated overnight at 37 °C with 5% CO2 . Cells were treated with E2 (1 μM) with and without SERMs (1 μM) for 6 days. Treatments were renewed after 3 days. Formation of ROS was determined as previously described (31), using CMH2DCFDA (10 μM) and 0.2 μg/mL Hoechst stain for visualization of nuclei.
Detection and measurement of 8-oxo-dG formation
MCF-10A cells were plated in 15 cm diameter dishes at a density of 2 × 106 cells/dish in estrogen free media. Cells were allowed to attach for 1 day and then were treated with 4-OHE2 (1 μM) with and without SERMs (1 μM) (DMA, Ral, or FDMA) for 72 h. 8-oxo-dG analysis was performed as described previously (17). The native dG was determined by HPLC (UV) scanning at 280 nm. 8-oxo-dG was detected by multiple reaction monitoring and collision-induced dissociation for the fragmentation pathway of m/z 284 → 168 and m/z 289 → 173 for (15N5)8-oxo-dG using positive ion electrospray. The amount of 8-oxo-dG formed per 106 of dG was plotted. Total 8-oxo-dG/106 of dG ratio for the 4-OHE2 treated sample was taken as 100% for the purpose of calculation.
Anchorage-independent growth assay
Anchorage independent colony formation cell transformation assay was performed as previously described (27). Spherical formation of >50 cells were taken as a colony. Number of colonies formed in each well were counted and represented as percentage colony efficiency ± SD. Percentage colony efficiency is calculated as the number of colonies formed per number of cells plated per well X100.
Immunoblotting
MCF-10A cells were treated with E2 (1 μM) in the presence and absence of SERMs (DMA, FDMA, Ral; 1 μM). Protein expression of CYP1B1 and 1A1 was analyzed using western blot experiments as previously described (27). Anti-CYP450 1B1 (Sigma; AV51761), Anti-CYP450 1A1 (Santa Cruz; sc-20772) and anti-β- actin (Cell signaling; #4967) antibodies were used as primary antibodies. Detoxification enzymes were also analyzed using anti-SULT1 (Santa Cruz,CA; sc-32928), anti-SULT1E1 (Santa Cruz, CA; sc-376009), anti-SULT1A1 (Santa Cruz, CA; sc-130883), anti-GSTpi (Cell signaling; #3369), anti-NQO1 (Santa Cruz; sc-32793), and anti COMT (Santa Cruz, CA; sc-25844) as primary antibodies. Antibodies were diluted in blocking solution (5% non fat milk in TBS with 0.1% tween 20). Blots were incubated with primary antibody overnight at 4 °C and with secondary antibody for 1 h at room temperature. Blots were visualized using chemiluminescence substrate (Thermo scientific, Rockford, IL). Imaging and analysis was done using FluroChem software (Cell Biosciences, Santa Clara, CA). Each protein band density was normalized to the respective β- actin band density and was represented as the relative protein expression. Three independent experiments were performed and results were represented as average ± SD.
RNA isolation and quantification of metabolizing enzyme gene transcripts
MCF-10A cell were plated at a density of 2 × 105 cells/ well in a 6 well plate and treated with E2 (1 μM) with and without SERMs (1 μM) for 24 h. Total RNA was isolated from cells using QIAShredder columns and QIAGEN RNeasy kit (Qiagen Inc., Valencia, CA) according to the manufacturer's protocol. Total RNA (1 μg) was used to synthesize cDNA using SuperScript III in a 20 μL reaction mixture according to manufacturer's protocol. qPCR analysis was done with respective primers. TaqMan FAM probes and primers (AB Applied Biosystems, Foster City, CA) were used for the gene analysis of SULT 1A1, SULT 1E1, and UGT 1A1 while human β-actin gene amplification was used as the internal control. Expression of the gene of interest was normalized to the internal control and fold change in gene expression was calculated. Three independent experiments were performed in duplicates and the data was represented as an average ± SD.
Enzyme activity assays
Inhibition of CYP450 1B1 activity was analyzed using ethoxyresorufin O-dealkylase (EROD) assay as previously described (27). Inhibition of COMT was assayed by adaptation of a literature method (32). Recombinant COMT (10 μg/mL) was incubated in Tris (10 mM, pH 7.4), MgCl2 (1 mM), DTT (1 μM), S-(5'-adenosyl)-L-methionine (500 nM) with or without Ral, Baze, or DMA (1 μM) at 37 °C for 5 min prior to initiation of reaction by addition of 6,7-dihydroxycoumarin (5 μM). Reaction was monitored by fluorescence ( λex = 355nm, λem = 460 nm).
Statistical analysis
Three independent metabolism experiments were performed in triplicates and the data was represented as average ± SD. The statistical analysis of results consisted of t-test or ANOVA using GraphPad Prism version 5 for Windows.
Results
DMA, Ral, and Baze, but not 4-OHTam, inhibit estrogen metabolism in MCF-10A cells
Analysis of E1 methoxy ethers is a useful indirect measurement of the formation of catechol metabolites in the presence of SERMs, since: (1) in MCF-10A cells, catechol estrogens are largely metabolized to methoxyethers that cannot themselves be directly converted to quinones; and (2) SERMs do not inhibit COMT activity (Supplementary Figure S1). After 6 days of E2 treatment, higher amounts of E1 relative to E2 metabolites, and relatively higher amounts of the 2-MeOE1 isomer were observed in all treatments (Supplementary Fig. S2).
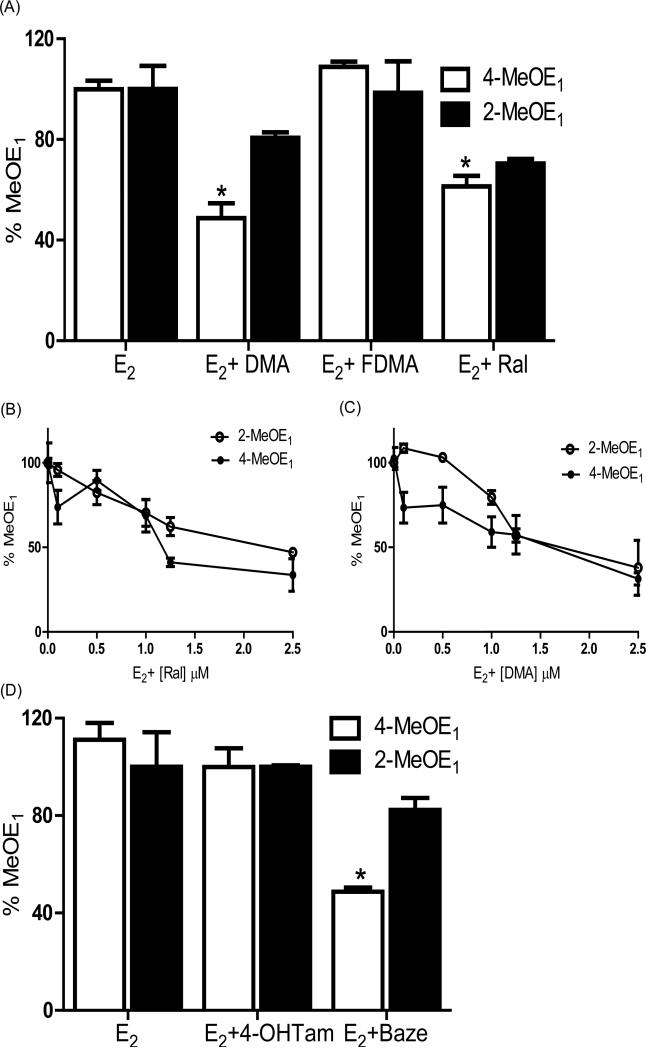
MCF-10A cells incubated with E2 (1 μM) were treated with vehicle or SERMs (1 μM) for 6 days and the formation of 4-MeOE1 and 2-MeOE1 was analyzed by LC/MS-MS, which provides a measure of catechol estrogen formation (Figure 2) (17, 27). Attenuation of MeOE1 formation with DMA and Ral reached significance for 4-MeOE1 (p < 0.05), whereas FDMA was without effect (Figure 2A). For Ral and DMA, the reduction in catechol ether formation was found to be concentration-dependent (Figures 2B, 2C). The effects on estrogen metabolism of the clinical SERMs, Baze and Tam, were also studied. No significant effect on metabolite formation was observed with 4-OHTam, the active metabolite of Tam, whereas Baze showed significant inhibition of 4-MeOE1 formation (p < 0.05; Figure 2D).
Figure 2.
(A) DMA and Ral significantly inhibit estrone methyl ether (MeOE1) formation in MCF-10A cells. MCF-10A cells were treated for 6 days with E2 (1 μM) in the presence and absence of SERMs (1 μM). Cell media was analyzed for MeOE1 and 4-MeOE1 formation using LC-MS/MS and an internal standard. “% MeOE1” was normalized to 100% representing cells treated with E2 alone. Attenuation of 4-MeOE1 formation with DMA or Ral co-treatment was significant; FDMA had no effect. Each data point represents an average of three independent experiments ± SD; * p < 0.05. (B), DMA and (C) Ral showed a dose dependent inhibition in both 4-MeOE1 and 2-MeOE1 formation in MCF-10A cells. Each data point represents an average of three independent experiments ± SD. (D) Modulation of MeOE1 formation by Baze or 4-OHTam co-treatment. Each data point represents an average of three independent experiments ± SD; * p < 0.05.
DMA, Ral, and Baze attenuate estrogen-induced ROS in MCF-10A cells
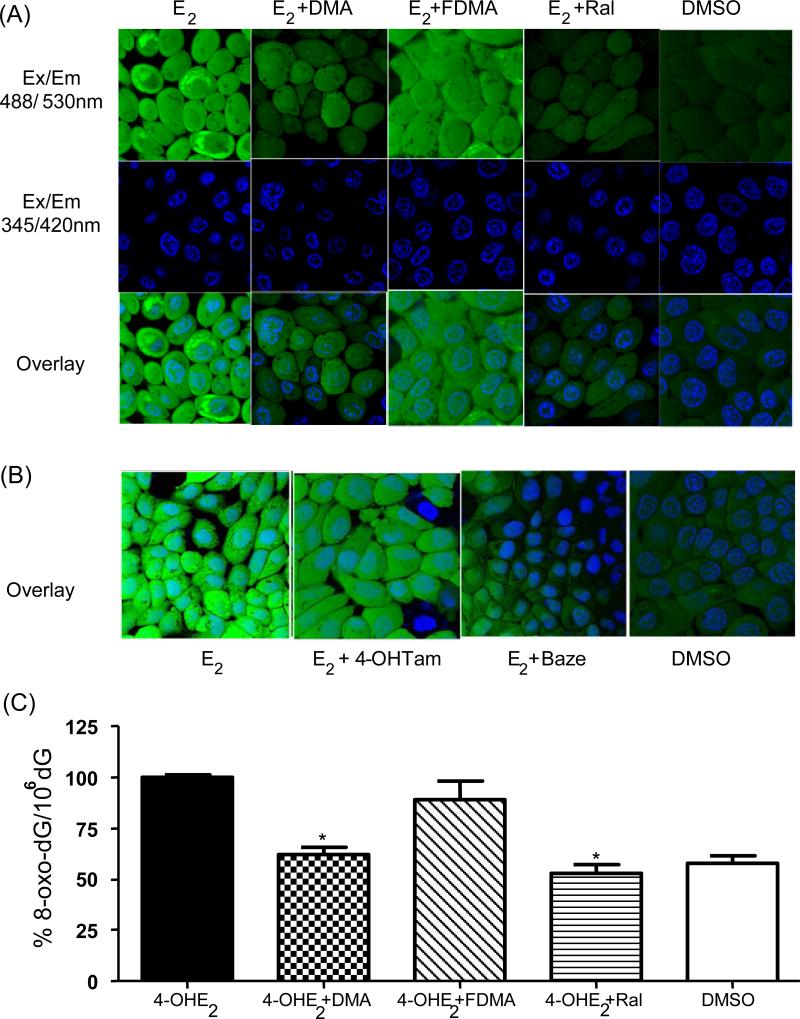
MCF-10A cells incubated with E2 for 6 days and treated with the reporter dye CMH2DCFDA, showed increased ROS levels compared to the DMSO vehicle control (Figure 3A). In ER(+) cells, localization of ER in the nucleus has been reported to produce nuclear ROS localization (31, 33); however, in MCF-10A cells, localization was not observed, compatible with the lack of function of ERα as a nuclear transcription factor in this cell line.
Figure 3.
(A) E2-induced formation of reactive oxygen species (ROS) in MCF-10A cells was inhibited by DMA and Ral, while FDMA had little effect. MCF-10A cells were treated with E2 in the presence and absence of SERMs for 6 days and ROS were labeled with CM-H2-DCFDA (10 μM) for 1 h. Nuclei were labeled with Hochest nuclear dye and DMSO (0.01%) treated cells were taken as the vehicle control. Live cells were imaged using confocal microscope META 510: green (Ex/Em; 488/530 nm) shows oxidized DCF-DA; blue (Ex/Em; 345/420 nm) shows nuclei. (B) E2-induced ROS formation in MCF-10A cells was attenuated with Baze co-treatment while 4-OHTam had little effect. (C) DMA and Ral significantly inhibit 4-OHE2 induced 8-oxodG generation in MCF-10A cells while FDMA was without significant effect. MCF-10A cells were treated with 4-OHE2 (1 μM) for 3 days and DNA was extracted and hydrolyzed to detect 8-oxo-dG using LC/MS-MS. All treatment groups used DMSO as vehicle. Each data point represents an average of three independent experiments ± SD; * p < 0.05.
E2-induced ROS formation was attenuated in cells co-treated with either DMA or Ral; however, there was no significant effect on the formation of ROS with FDMA co-treatment (Figure 3A). Baze and 4-OHTam were also tested for their effect on E2-induced ROS formation in MCF-10A cells: no significant effect was observed on 4-OHTam treatment; however, Baze attenuated ROS formation (Figure 3B).
DMA and Ral significantly attenuate 4-OHE2 induced 8-oxo-dG formation
Measurement of 8-oxo-dG is routinely used to determine the level of oxidative DNA damage in cells and in vivo (34). After 3 days treatment of MCF-10A cells with E2, formation of 8-oxo-dG did not reach significance relative to DMSO control (data not shown), therefore, MCF-10A cells were treated directly with the catechol estrogen metabolite, 4-OHE2 (1 μM), for 3 days revealing a significant increase in 8-oxo-dG relative to DMSO control (p < 0.001). Co-treatment with either DMA or Ral significantly reduced (p < 0.05) 8-oxo-dG levels induced by 4-OHE2. Co-administration of FDMA was again without effect (Figure 3C).
DMA and Ral do not decrease CYP450 expression or activity
CYP450 enzymes mediate estrogen-induced chemical carcinogenesis by catalyzing catechol estrogen formation (Figure 1). CYP450 expression was analyzed by immunoblotting after treatment of MCF-10A cells with E2 in the presence or absence of SERMs. showing no effect of SERM cotreatment on CYP450 levels (Supplementary Fig. S3). Measurement of CYP450 1B1 activity using the EROD assay revealed the expected inhibition by SERMs at very high concentrations, but not at the 1 μM concentration applied to cells (Supplementary Fig. S4).
DMA and Ral detoxify estrogen metabolites via action on Phase 2 enzymes
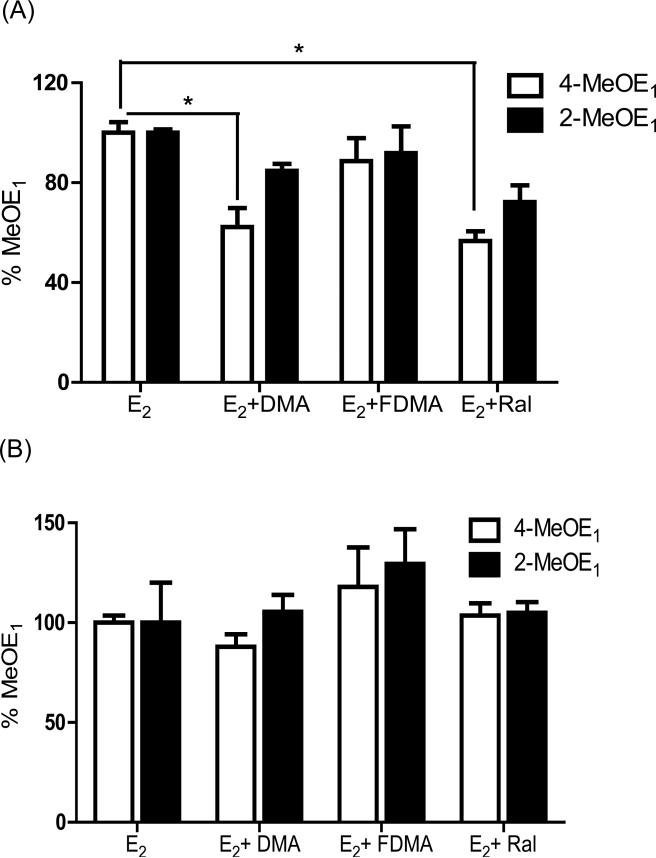
Sulfation and glucuronidation play key roles in conjugative detoxification, therefore, levels of estrogen metabolites in cell media were measured in the presence of a sulfatase/β-glucuronidase cocktail that causes enzymatic hydrolysis of conjugates. The attenuation of 4-MeOE1 and 2-MeOE1 formation by SERMs was completely lost under these conditions (Figure 4), leading to the conclusion that conjugative metabolism is responsible for the attenuation of catechol estrogen metabolite formation caused by SERM co-treatment.
Figure 4.
Attenuation of MeOE1 formation on co-treatment with Ral or DMA (A); was lost after addition of β- glucuronidase and sulfatase to supernatants (B). Hydrolysis of glucuronate and sulfate groups from estrone metabolites negates the observed effect of SERM co-treatment on relative MeOE1 formation in MCF-10A cells treated with E2 (1 μM). After 6 days treatment, cell media was collected and divided into two portions where one portion was incubated with β-glucuronidase and sulfatase at 37 °C overnight and the second portion represented the control sample. Analysis was by LC/MS-MS measured using an internal standard. Each data point represents an average of three independent experiments ± SD; * p < 0.05.
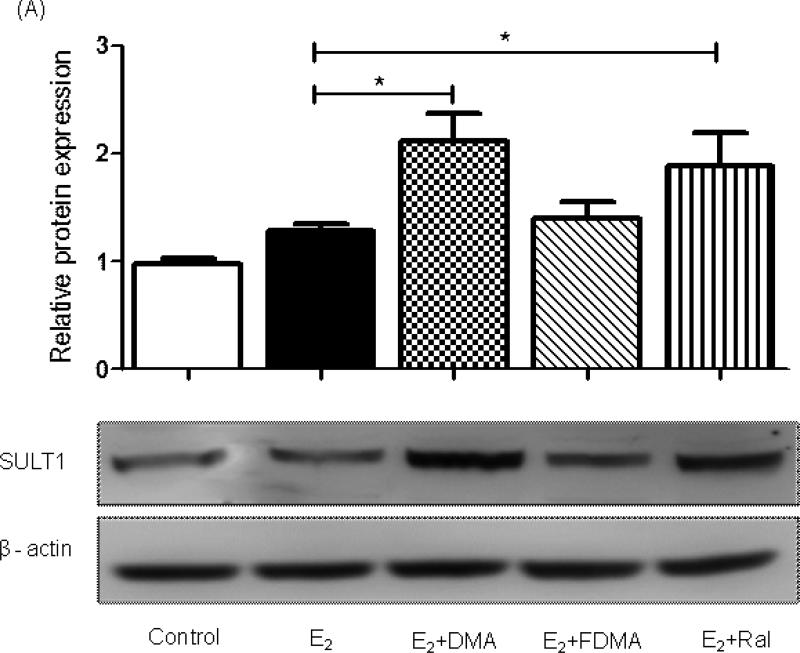
Extending this observation, co-treatment of MCF-10A cells with E2 and either DMA or Ral significantly elevated immunoreactivity of SULT1 family enzymes (Figure 5A). Further analysis indicated that expression of SULT 1E1 was induced by co-treatment with DMA or Ral (Supplementary Fig. S5) but that SULT 1A1 expression was not significantly changed (Supplementary Fig. S5). E2 itself did not modulate SULT1 expression and once again, FDMA was unable to mimic the effects of Ral and DMA (Figure 5A). Reduction of quinones by NQO1 is able to maintain a reducing cellular environment, unless this activity contributes to redox cycling (11, 22) (Figure 1). The reduction in NQO1 expression after treatment of MCF-10A cells with E2, was negated or reversed by co-treatment with SERMs (Supplementary Fig. S4). Similar analyses of COMT and GST-P1 expression showed no effect from co-treatment with SERMs (Supplementary Fig. S5). The lack of sensitivity of COMT to E2 and drug treatments further supports measurements of MeOE1 as reflective of catechol estrogen formation.
Figure 5.
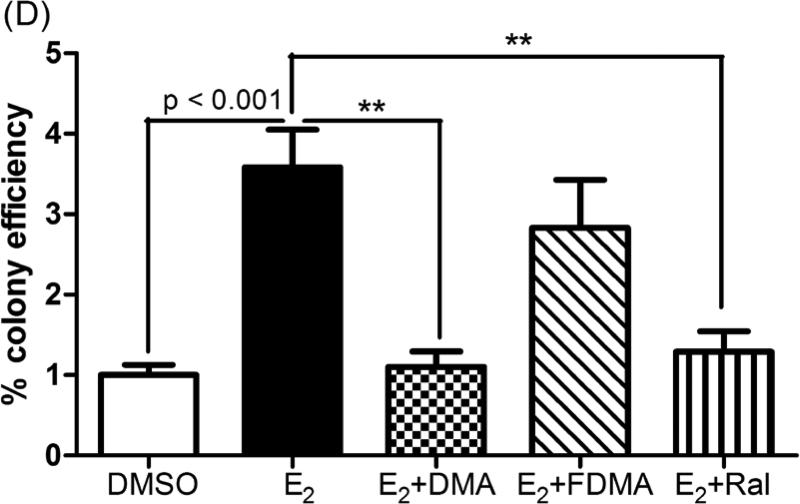
(A) A significant induction in SULT 1 enzyme expression was observed by immunoblotting on co-treatment of MCF-10A cells with DMA or Ral. Relative protein amounts were determined by densitometric analysis of SULT1 protein after western blot, loading with 30 μg of total protein. Each treatment was normalized to the loading and transferring control, β actin. Each data point represents an average of three independent experiments in duplicate ± SD; * p < 0.05. (B) Gene transcription of SULT 1E1 was significantly induced by DMA and Ral while FDMA had no effect compare to E2 treatment alone. (C) Gene transcription of UGT 1A1 was significantly induced co-treatment with DMA or Ral as measured by qPCR after isolating RNA from 24 h treated MCF-10A cells. Each data point represents an average of three independent experiments ± SD; * p < 0.05, ** p < 0.01. (D) Co-treatment with DMA or Ral significantly inhibited E2-induced anchorage-independent colony growth of MCF-10A cells in soft agar, while FDMA co-treatment had little effect. Cells were treated twice a week, in the presence and absence of SERMs, over the course of 3 weeks. DMSO (0.01%) was used as the vehicle control in the experiments in the absence of E2 treatment. Cells were plated on soft agar and maintained for 3 weeks. Relative colony efficiency is calculated by dividing the number of colonies counted in a well by the number of cells plated in each well, normalized to DMSO vehicle. Data shows mean from three independent experiments ± SD: ** p < 0.005.
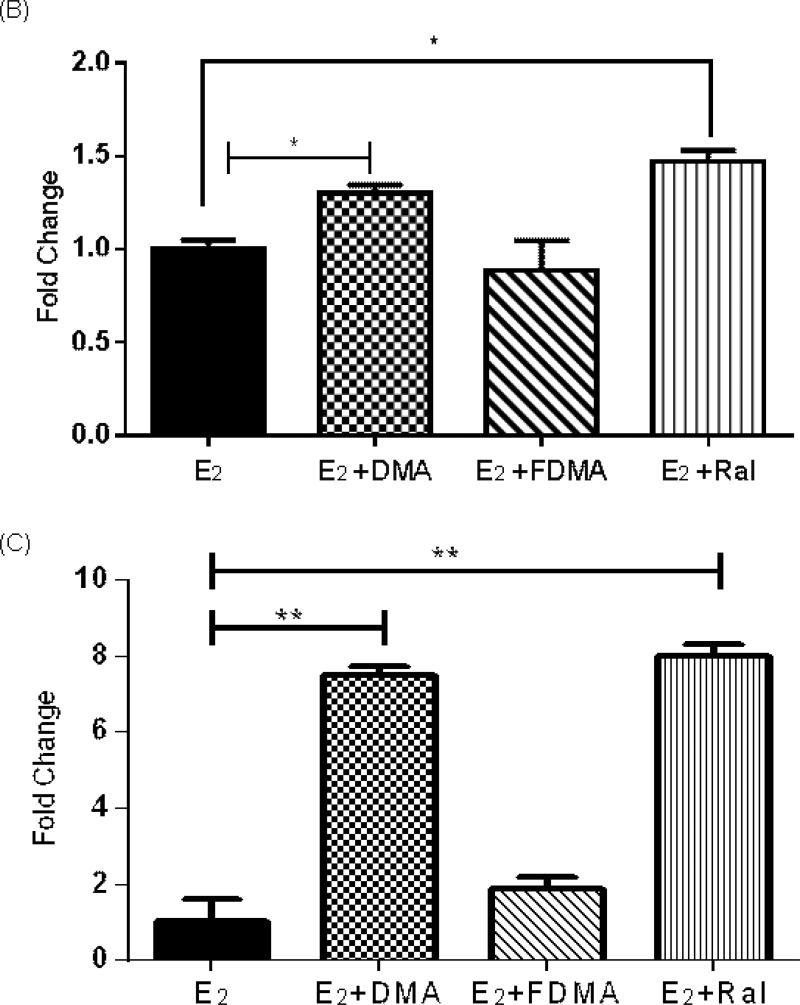
Since immunoblotting showed induction of SULT 1 family proteins after co-treatment with DMA or Ral, qPCR experiments were conducted to examine the effect of SERMs on SULT1E1 and SULT1A1. A significant increase in gene transcription of SULT1E1 was observed with co-treatment of DMA and Ral (p < 0.05), while the effect of FDMA was not significant (Figure 5B). There was an induction of SULT1A1 gene transcription in E2 incubations, which was not significantly perturbed by SERM co-treatment (Supplementary Fig. S5). Induction of UGT 1A1 (p < 0.05) was observed with both DMA and Ral co-treatment, while the effect of FDMA was not significant (Figure 5C). Transcription of SULT1E1, in response to SERM co-treatment, mirrored the observations on protein expression.
DMA and Ral significantly inhibit E2-induced anchorage-independent colony formation
Upon exposure to chemical carcinogens, MCF-10 cells can be transformed into a malignant phenotype reflected by formation of anchorage-independent colonies (35, 36). MCF-10A cells treated with E2 (1 μM) for 3 weeks underwent malignant transformation as shown by formation of colonies in soft agar (27). E2-induced colony formation was significantly inhibited by co-treatment with DMA and Ral (p < 0.05), whereas FDMA was without effect (Figure 5D).
Modulation of estrogen metabolism in MCF-10A cells is not mediated by classical ER
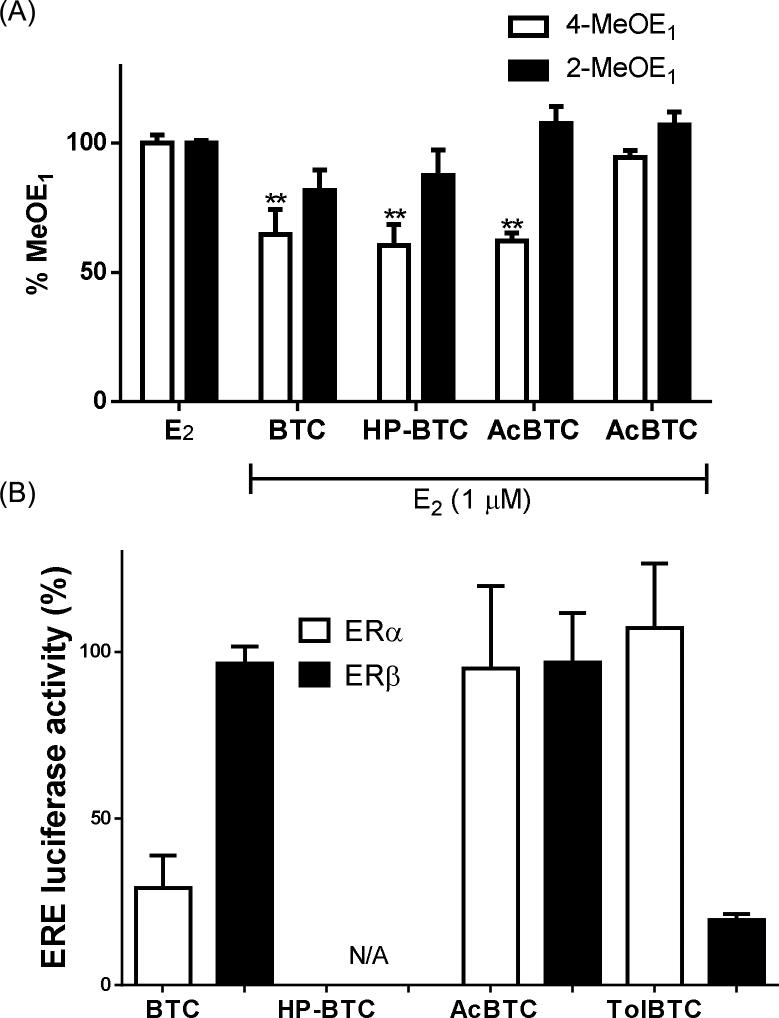
MCF-10A cells are formally considered as ER(-), since estrogen does not induce proliferation; however, the presence of ER protein and mRNA has been determined in MCF-10A cells (37-41). Since the primary biological target of SERMs is ER, it was essential to determine if classical ER signaling via these proximal receptors was causal in modulation of oxidative metabolism to produce catechol estrogens. We therefore chose to study the effects on E2 metabolism of analogues of DMA (BTC, HP-BTC, AcBTC, TolBTC) with varied activity at ERα and ERβ (42). The formation of 4-MeOE1 was measured in E2-treated MCF-10A cells co-treated with DMA analogues: TolBTC was without effect; whereas BTC, HP-BTC, and AcBTC significantly reduced oxidative metabolite formation (Figure 6A). Using ERE-luciferase reporters, full concentration-response curves were obtained for DMA analogues in MCF-7 and in MDA-MB-231:β41 cells, to determine EC50 for classical ERα and ERβ signaling, respectively (Supplementary Fig. S6). The relative luciferase activity for the DMA analogues illustrates that BTC is an ERβ selective agonist, TolBTC is an ERα selective agonist, Ac-BTC is a non-selective agonist, and HP-BTC was observed to be an antagonist (Figure 6B). Ral, DMA, FDMA, 4-OHTam, and Baze have been extensively profiled by ourselves and others as classical ERα antagonists in mammary epithelial cell lines (28, 43). The collected classical ERα and ERβ activity data on DMA analogues and SERMs shows no correlation with observed effects on accumulation of catechol estrogen metabolites in MCF-10A cells.
Figure 6.
(A) Co-treatment with BTC, HP-BTC, or AcBTC significantly inhibited oxidative estrogen metabolism in E2-treated MCF-10A cells as shown by measurement of MeOE1 after 6 days. Normalization of %MeOE1 with respect to cells treated with 100%E2 alone is described fully in the text. Each data point represents an average of three independent experiments in duplicate ± SD: ** p < 0.01. (B) Classical ER/ERE signaling measured using a pERE-luciferase reporter after treatment of cells for 24 h with BTC, HP-BTC, AcBTC, or TolBTC: in MCF-7 cells to obtain data for ERα; and in MDA-MB-231:β41 cells to test for ERβ-mediated activity. Data represent an average of three independent experiments performed in duplicates ± SD.
Discussion
SERMs are used in the treatment and prevention of postmenopausal osteoporosis (44) and also in primary and secondary prevention of ER(+) breast cancer, with STAR (Study of Tamoxifen and Ral) and IBIS2 (International Breast cancer Intervention Study 2) reporting data on primary chemoprevention (45). In light of the clinical use of SERMs and both the current and potential use in breast cancer chemoprevention, the present study was designed to determine the effect of SERMs on estrogen-induced chemical carcinogenesis, a pathway that is independent of the formal ER status of cells and tissues. We hypothesized that modulation of oxidative estrogen metabolism in mammary epithelial cells by SERMs would influence the estrogen-induced malignant transformation of these cells, and be of relevance to chemical carcinogenesis.
When MCF-10A non-tumorigenic breast epithelial cells, in the presence of E2, were co-treated with selected clinical or preclinical SERMs, formation of MeOE1 catechol estrogen metabolites was significantly attenuated. The attenuated metabolism correlated with the effect of Ral and DMA in preventing E2-induced malignant transformation of MCF-10A cells (Figure 5D), representing the first evidence that modulation by SERMs of estrogen metabolism in mammary cells attenuates malignant transformation. In contrast to DMA and Ral, FDMA, an analogue of DMA, with similar affinity and potency at ER to Ral and DMA (46, 47), caused no significant attenuation of estrogen metabolism and did not inhibit malignant transformation. The clinical SERM, Baze, was also observed to inhibit formation of oxidative estrogen metabolites; whereas, Tam, had no effect on metabolism (Figure 2D). Therefore, attenuation of estrogen oxidative metabolism is not a feature common to the entire SERM drug class; however, where studied, attenuated metabolism correlated with inhibition of malignant transformation.
Both estrogen-induced ROS formation and formation of 8-oxo-dG are indicators of oxidative stress and possible genotoxicity leading to carcinogenesis (34, 48, 49). Exposure of breast epithelial cells to catechol estrogen metabolites is associated with ROS formation (12, 50). In the present study, we observed that exposure to E2 for 6 days significantly increased ROS in MCF-10A cells (Figure 3A). There was a clear correlation between ROS formation and metabolism to catechol estrogens with all clinical and pre-clinical SERMs tested. Treatment of MCF-10A cells directly with the catechol estrogen metabolite, 4-OHE2, gave a significant increase in 8-oxo-dG after 3 days, compatible with induction of oxidative DNA damage by this carcinogenic metabolite (48, 51). Co-treatment of cells with DMA and Ral, but not FDMA, led to inhibition of both estrogen-induced ROS and 8-oxo-dG formation (Figure 3).
The MCF-10A cell line represents a model system to evaluate estrogen metabolism and malignant transformation in vitro (21). Previous studies have shown that both MCF-10A and MCF-10F cells can be transformed into a malignant phenotype upon exposure to E2 and 4-OHE2 (27, 52) leading to the formation of anchorage independent colonies in semi solid media. It has been previously reported that a botanical extract of hops (Humulus lupulus), could significantly reduce estrogen induced malignant transformation in MCF-10A cells via attenuation of oxidative estrogen metabolism (27). Malignant transformation of MCF-10F cells, induced by a combination of estrogen and TCDD, was also reported to be inhibited by attenuation of estrogen metabolism, on resveratrol co-treatment (19, 53). In the present study, DMA and Ral significantly inhibited E2-induced malignant transformation by attenuation of catechol estrogen metabolite formation and concomitant reduction in levels of ROS and 8-oxodG.
Transformation of normal breast epithelial cells into a malignant phenotype, measured by formation of anchorage-independent colonies in semi-solid media, is dependent upon oxidative hydroxylation of E2 to a catechol metabolite. Estrogen metabolism can be modulated either via: i) downregulating or inhibiting CYP450 enzymes and thereby reducing the formation of catechols and quinones; or ii) induction of Phase 2 enzymes that “detoxify” these catechol metabolites by conjugation and elimination. Hops extract and other agents have been reported to inhibit the expression of CYP450 1B1 in human breast epithelial cells (27, 53, 54); however, no evidence for regulation or inhibition of CYP450 1A1 or CYP450 1B1 by Ral or DMA was observed at the concentrations used in MCF-10A cell cultures.
Phase 2 conjugative enzyme activity has been reported to correlate with malignant transformation in vitro, tumorigenesis in vivo, and breast cancer risk in human subjects. SULTs play a major role in hepatic and extra-hepatic detoxification of xenobiotics and other toxic metabolites. It has been reported that SULT 1E1 and SULT 2B1 are responsible for detoxification of estrogenic catechols via sulfation (40). In MCF-10A cells, one paper reported that both SULTs were equally expressed at the mRNA level and more highly so than in breast cancer cells such as T47D, SKBR3, and MDA-MB-231 (40). Another group reported expression of SULT 1E1 mRNA alone in MCF-10A cells and observed both epigenetic regulation of SULT 1E1 mRNA and repression in transformed MCF-10A-derived cells (39). There is some evidence to suggest that SULT 1E1 gene transcription is mediated via the aryl hydrocarbon receptor in MCF-10A cells (55). In addition, there may be an association between breast cancer and genetic polymorphisms in human UGT1A1, another key mediator of conjugative metabolism (56).
The expression of SULT and UGT was assayed with and without SERM co-treatment in E2-treated MCF-10A cells: SULT 1E1 expression was induced by DMA and Ral. UGT 1A1 was also induced by DMA and Ral co-treatment, however, the expression of UGT 1A1 was low in MCF-10A cells when tested by immunoblotting. The inhibitory effects of DMA and Ral on MeOE1 formation were lost when supernatants were treated with sulfatase/glucuronidase (Figure 4). Future studies of the role of glucuronidation in detoxification will interrogate formation of the hydrophilic glucuronate and sulfate metabolites. However, the combined observations support induction of Phase 2 metabolism, and particularly SULT 1E1 expression, as a mechanism of detoxification of carcinogenic estrogen metabolites by selected SERMs in breast epithelial cells.
FDMA, proved to be an excellent probe of mechanism, because of the SERMs studied, FDMA alone did not attenuate metabolite levels, nor upregulate SULT 1E1, nor inhibit both ROS and 8-oxo-dG formation. Clear trends were observed in reduction of the 2-MeOE1 metabolite by DMA and Ral, although significance was not reached. Several lines of evidence strongly suggest that the 4-OH catechol estrogen is the proximal carcinogen in estrogen chemical carcinogenesis [13-17]. It is possible that in the model system under study malignant transformation can be elicited by both isomeric catechol estrogens; however, more detailed study would be needed.
It was important to confirm that the observed expression of SULT 1E1, attenuation of estrogen metabolism, and inhibition of malignant transformation, was not mediated via ligand binding to ER and resulting classical, ERE-mediated transcription. The lack of effect observed for FDMA and 4HO-Tam, both high affinity ER ligands, supports this assertion. Furthermore, estrogen metabolism was measured in the presence of DMA analogues that manifest varied selectivity for ERα and ERβ and agonist/antagonist activity at ER: no correlation was observed between attenuation of estrogen metabolism and ER/ERE signaling.
Interestingly, the phytoestrogen genistein has been the subject of two recent studies in MCF-10A cells, implicating independently upregulation of detoxification enzymes (57) and of PTEN (58) in mediating chemoprevention. The cause was speculatively attributed to ligand binding to ERβ or the G-protein coupled receptor, GPR30 (GPER) (59). GPR30 mediates many non-classical, extranuclear actions of estrogens and antiestrogens, including the actions of Ral, DMA, and DMA analogues (42). However, several SERMs are able to act as phenolic antioxidants and to activate stress response via Nrf2 and the antioxidant response element (ARE) (60-63), therefore ER-independent pathways are known that might regulate function in MCF-10A cells by DMA, Ral, and Baze.
The present study demonstrates that clinical SERMs can attenuate estrogen chemical carcinogenesis by modulating oxidative estrogen metabolism. Treatment of human breast epithelial cells with the clinical SERMs, Ral, DMA, and Baze, but not 4-OHTam, led to inhibition of oxidative estrogen metabolism. Attenuated oxidative metabolism and lower levels of ROS were correlated with inhibition of E2-induced malignant transformation. The mechanism of inhibition by Ral and DMA was shown to be detoxification of genotoxic estrogen metabolite accumulation mediated via upregulation of SULT 1E1. Further studies are underway to identify the proximal receptor for these SERMs and to extend studies to animal models.
Supplementary Material
Acknowledgment
We thank Maitrayee Bose and Ping Yao for their technical support.
Financial support: Support for this work was provided by NIH grants CA79870 (J.L.Bolton) and CA102590 (G.R.J. Thatcher).
Abbreviations
- AhR
Aryl hydrocarbon receptor
- Baze
Bazedoxifene
- COMT
catechol O-methyl transferase
- DMA
Desmethylarzoxifene
- E2
17β-estradiol
- ER
estrogen receptor
- FDMA
4’-floro DMA
- GST
glutathione-S-transferase
- HRT
hormone replacement therapy
- 2-OHE1
2-hydroxyestrone
- 4-OHE1
4-hydroxyestrone
- 2-OHE2
2-hydroxyestradiol
- 4-OHE2
4-hydroxyestradiol
- 4-OHTam
4-hydroxytamoxifen
- LC/MS-MS
liquid chromatography-tandem mass spectrometry
- 2-MeOE1
2-methoxyestrone
- 4-MeOE1
4-methoxyestrone
- 2-MeOE2
2-methoxyestradiol, 4-MeOE2, 4-methoxyestradiol
- NQO1
NAD(P)H:quinone oxidoreductase
- Ral
Raloxifene
- ROS
reactive oxygen species
- SERMs
selective estrogen receptor modulators
- SULT
sulfotransferase
- Tam
Tamoxifen
- UGT
UDP-glucuronosyltransferase
References
- 1.Russo J, Russo IH. Biological and molecular bases of mammary carcinogenesis. Lab Invest. 1987;57:112–37. [PubMed] [Google Scholar]
- 2.Feigelson HS, Henderson BE. Estrogens and breast cancer. Carcinogenesis. 1996;17:2279–84. doi: 10.1093/carcin/17.11.2279. [DOI] [PubMed] [Google Scholar]
- 3.Henderson BE, Feigelson HS. Hormonal carcinogenesis. Carcinogenesis. 2000;21:427–33. doi: 10.1093/carcin/21.3.427. [DOI] [PubMed] [Google Scholar]
- 4.Yager JD, Davidson NE. Estrogen Carcinogenesis in Breast Cancer. N Engl J Med. 2006;354:270–82. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- 5.Bolton JL, Pisha E, Zhang F, Qiu S. Role of quinoids in estrogen carcinogenesis. Chem Res Toxicol. 1998;11:1113–27. doi: 10.1021/tx9801007. [DOI] [PubMed] [Google Scholar]
- 6.Bolton JL, Thatcher GRJ. Potential mechanisms of estrogen quinone carcinogenesis. Chem Res Toxicol. 2008;21:93–101. doi: 10.1021/tx700191p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavalieri EL, Stack DE, Devanesan PD, Todorovic R, Dwivedy I, Higginbotham S, et al. Molecular origin of cancer: Catechol estrogen-3,4-quinones as endogenous tumor initiators. Proc Natl Acad Sci U S A. 1997;94:10937–42. doi: 10.1073/pnas.94.20.10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iida T, Furuta A, Kawashima M, Nishida J, Nakabeppu Y, Iwaki T. Accumulation of 8-oxo-2′-deoxyguanosine and increased expression of hMTH1 protein in brain tumors. Neuro Oncol. 2001;3:73–81. doi: 10.1093/neuonc/3.2.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roszkowski K, Jozwicki W, Blaszczyk P, Mucha-Malecka A, Siomek A. Oxidative damage DNA: 8-oxoGua and 8-oxodG as molecular markers of cancer. Med Sci Monit. 2011;17:CR329–33. doi: 10.12659/MSM.881805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kryston TB, Georgiev AB, Pissis P, Georgakilas AG. Role of oxidative stress and DNA damage in human carcinogenesis. Mutat Res. 2011;711:193–201. doi: 10.1016/j.mrfmmm.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z, Chandrasena ER, Yuan Y, Peng KW, van Breemen RB, Thatcher GR, et al. Redox cycling of catechol estrogens generating apurinic/apyrimidinic sites and 8-oxo-deoxyguanosine via reactive oxygen species differentiates equine and human estrogens. Chem Res Toxicol. 2010;23:1365–73. doi: 10.1021/tx1001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fussell KC, Udasin RG, Smith PJS, Gallo MA, Laskin JD. Catechol metabolites of endogenous estrogens induce redox cycling and generate reactive oxygen species in breast epithelial cells. Carcinogenesis. 2011;32:1285–93. doi: 10.1093/carcin/bgr109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li JJ, Li SA. Estrogen carcinogenesis in Syrian hamster tissues: role of metabolism. Fed Proc. 1987;46:1858–63. [PubMed] [Google Scholar]
- 14.Zhu BT, Bui QD, Weisz J, Liehr JG. Conversion of estrone to 2- and 4-hydroxyestrone by hamster kidney and liver microsomes: implications for the mechanism of estrogen-induced carcinogenesis. Endocrinology. 1994;135:1772–9. doi: 10.1210/endo.135.5.7956900. [DOI] [PubMed] [Google Scholar]
- 15.Han X, Liehr JG. Microsome-mediated 8-hydroxylation of guanine bases of DNA by steroid estrogens: correlation of DNA damage by free radicals with metabolic activation to quinones. Carcinogenesis. 1995;16:2571–4. doi: 10.1093/carcin/16.10.2571. [DOI] [PubMed] [Google Scholar]
- 16.Zhao Z, Kosinska W, Khmelnitsky M, Cavalieri EL, Rogan EG, Chakravarti D, et al. Mutagenic activity of 4-hydroxyestradiol, but not 2-hydroxyestradiol, in BB rat2 embryonic cells, and the mutational spectrum of 4-hydroxyestradiol. Chem Res Toxicol. 2006;19:475–9. doi: 10.1021/tx0502645. [DOI] [PubMed] [Google Scholar]
- 17.Kastrati I, Edirisinghe PD, Hemachandra LP, Chandrasena ER, Choi J, Wang YT, et al. Raloxifene and desmethylarzoxifene block estrogen-induced malignant transformation of human breast epithelial cells. PLoS One. 2011;6:e27876. doi: 10.1371/journal.pone.0027876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rogan EG, Badawi AF, Devanesan PD, Meza JL, Edney JA, West WW, et al. Relative imbalances in estrogen metabolism and conjugation in breast tissue of women with carcinoma: potential biomarkers of susceptibility to cancer. Carcinogenesis. 2003;24:697–702. doi: 10.1093/carcin/bgg004. [DOI] [PubMed] [Google Scholar]
- 19.Lu F, Zahid M, Wang C, Saeed M, Cavalieri EL, Rogan EG. Resveratrol prevents estrogen-DNA adduct formation and neoplastic transformation in MCF-10F cells. Cancer Prev Res (Phila) 2008;1:135–45. doi: 10.1158/1940-6207.CAPR-08-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soule HD, Maloney TM, Wolman SR, Peterson WD, Brenz R, McGrath CM, et al. Isolation and Characterization of a Spontaneously Immortalized Human Breast Epithelial Cell Line, MCF-10. Cancer Res. 1990;50:6075–86. [PubMed] [Google Scholar]
- 21.Russo J, Hasan Lareef M, Balogh G, Guo S, Russo IH. Estrogen and its metabolites are carcinogenic agents in human breast epithelial cells. J Steroid Biochem Mol Biol. 2003;87:1–25. doi: 10.1016/s0960-0760(03)00390-x. [DOI] [PubMed] [Google Scholar]
- 22.Chandrasena RE, Edirisinghe PD, Bolton JL, Thatcher GRJ. Problematic detoxification of estrogen quinones by NAD(P)H-dependent quinone oxidoreductase and glutathione-S-transferase. Chem Res Toxicol. 2008 doi: 10.1021/tx8000797. [DOI] [PubMed] [Google Scholar]
- 23.Gaikwad NW, Rogan EG, Cavalieri EL. Evidence from ESI-MS for NQO1-catalyzed reduction of estrogen ortho-quinones. Free Radical Bio Med. 2007;43:1289–98. doi: 10.1016/j.freeradbiomed.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buckley DB, Klaassen CD. Tissue- and gender-specific mRNA expression of UDP-glucuronosyltransferases (UGTs) in mice. Drug Metab Dispos. 2007;35:121–7. doi: 10.1124/dmd.106.012070. [DOI] [PubMed] [Google Scholar]
- 25.Spink BC, Katz BH, Hussain MM, Pang S, Connor SP, Aldous KM, et al. SULT1A1 catalyzes 2-methoxyestradiol sulfonation in MCF-7 breast cancer cells. Carcinogenesis. 2000;21:1947–57. doi: 10.1093/carcin/21.11.1947. [DOI] [PubMed] [Google Scholar]
- 26.Tamura HO, Taniguchi K, Hayashi E, Hiyoshi Y, Nagai F. Expression profiling of sulfotransferases in human cell lines derived from extra-hepatic tissues. Biol Pharm Bull. 2001;24:1258–62. doi: 10.1248/bpb.24.1258. [DOI] [PubMed] [Google Scholar]
- 27.Hemachandra LP, Madhubhani P, Chandrasena R, Esala P, Chen SN, Main M, et al. Hops (Humulus lupulus) Inhibits Oxidative Estrogen Metabolism and Estrogen-Induced Malignant Transformation in Human Mammary Epithelial cells (MCF-10A). Cancer Prev Res (Phila) 2012;5:73–81. doi: 10.1158/1940-6207.CAPR-11-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Overk CR, Peng KW, Asghodom RT, Kastrati I, Lantvit DD, Qin Z, et al. Structure-activity relationships for a family of benzothiophene selective estrogen receptor modulators including raloxifene and arzoxifene. ChemMedChem. 2007;2:1520–6. doi: 10.1002/cmdc.200700104. [DOI] [PubMed] [Google Scholar]
- 29.Tonetti DA, Rubenstein R, DeLeon M, Zhao H, Pappas SG, Bentrem DJ, et al. Stable transfection of an estrogen receptor beta cDNA isoform into MDA-MB-231 breast cancer cells. J Steroid Biochem Mol Biol. 2003;87:47–55. doi: 10.1016/j.jsbmb.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 30.Xu X, Keefer LK, Ziegler RG, Veenstra TD. A liquid chromatography-mass spectrometry method for the quantitative analysis of urinary endogenous estrogen metabolites. Nat Protocols. 2007;2:1350–5. doi: 10.1038/nprot.2007.176. [DOI] [PubMed] [Google Scholar]
- 31.Wang Z, Wijewickrama GT, Peng KW, Dietz BM, Yuan L, van Breemen RB, et al. Estrogen Receptor {alpha} Enhances the Rate of Oxidative DNA Damage by Targeting an Equine Estrogen Catechol Metabolite to the Nucleus. J Biol Chem. 2009;284:8633–42. doi: 10.1074/jbc.M807860200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurkela M, Siiskonen A, Finel M, Tammela P, Taskinen J, Vuorela P. Microplate screening assay to identify inhibitors of human catechol-O-methyltransferase. Anal Biochem. 2004;331:198–200. doi: 10.1016/j.ab.2004.04.026. [DOI] [PubMed] [Google Scholar]
- 33.Peng KW, Wang H, Qin Z, Wijewickrama GT, Lu M, Wang Z, et al. Selective estrogen receptor modulator delivery of quinone warheads to DNA triggering apoptosis in breast cancer cells. ACS Chem Biol. 2009;4:1039–49. doi: 10.1021/cb9001848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loft S, Deng XS, Tuo J, Wellejus A, Sorensen M, Poulsen HE. Experimental study of oxidative DNA damage. Free Radic Res. 1998;29:525–39. doi: 10.1080/10715769800300571. [DOI] [PubMed] [Google Scholar]
- 35.Kim DW, Sovak MA, Zanieski G, Nonet G, Romieu-Mourez R, Lau AW, et al. Activation of NF-kappaB/Rel occurs early during neoplastic transformation of mammary cells. Carcinogenesis. 2000;21:871–9. doi: 10.1093/carcin/21.5.871. [DOI] [PubMed] [Google Scholar]
- 36.Calaf G, Russo J. Transformation of human breast epithelial cells by chemical carcinogens. Carcinogenesis. 1993;14:483–92. doi: 10.1093/carcin/14.3.483. [DOI] [PubMed] [Google Scholar]
- 37.Kastrati I, Edirisinghe PD, Wijewickrama GT, Thatcher GR. Estrogen-induced apoptosis of breast epithelial cells is blocked by NO/cGMP and mediated by extranuclear estrogen receptors. Endocrinology. 2010;151:5602–16. doi: 10.1210/en.2010-0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spink DC, Spink BC, Cao JQ, DePasquale JA, Pentecost BT, Fasco MJ, et al. Differential expression of CYP1A1 and CYP1B1 in human breast epithelial cells and breast tumor cells. Carcinogenesis. 1998;19:291–8. doi: 10.1093/carcin/19.2.291. [DOI] [PubMed] [Google Scholar]
- 39.Fu J, Weise AM, Falany JL, Falany CN, Thibodeau BJ, Miller FR, et al. Expression of estrogenicity genes in a lineage cell culture model of human breast cancer progression. Breast Cancer Res Treat. 2010;120:35–45. doi: 10.1007/s10549-009-0363-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hevir N, Trost N, Debeljak N, Rizner TL. Expression of estrogen and progesterone receptors and estrogen metabolizing enzymes in different breast cancer cell lines. Chem Biol Interact. 2011;191:206–16. doi: 10.1016/j.cbi.2010.12.013. [DOI] [PubMed] [Google Scholar]
- 41.Wang J, Gildea JJ, Yue W. Aromatase overexpression induces malignant changes in estrogen receptor alpha negative MCF-10A cells. Oncogene. 2013;32:5233–40. doi: 10.1038/onc.2012.558. [DOI] [PubMed] [Google Scholar]
- 42.Abdelhamid R, Luo J, Vandevrede L, Kundu I, Michalsen B, Litosh VA, et al. Benzothiophene Selective Estrogen Receptor Modulators Provide Neuroprotection by a novel GPR30-dependent Mechanism. ACS Chem Neurosci. 2011;2:256–68. doi: 10.1021/cn100106a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller CP, Collini MD, Tran BD, Harris HA, Kharode YP, Marzolf JT, et al. Design, synthesis, and preclinical characterization of novel, highly selective indole estrogens. J Med Chem. 2001;44:1654–7. doi: 10.1021/jm010086m. [DOI] [PubMed] [Google Scholar]
- 44.Komm BS, Chines AA. An update on selective estrogen receptor modulators for the prevention and treatment of osteoporosis. Maturitas. 2011 doi: 10.1016/j.maturitas.2011.11.018. [DOI] [PubMed] [Google Scholar]
- 45.Dutertre M, Smith CL. Molecular mechanisms of selective estrogen receptor modulator (SERM) action. J Pharmacol Exp Ther. 2000;295:431–7. [PubMed] [Google Scholar]
- 46.Liu H, Bolton JL, Thatcher GRJ. Chemical modification modulates estrogenic activity, oxidative reactivity, and metabolic stability in 4′F-DMA, a new benzothiophene selective estrogen receptor modulator. Chem Res Toxicol. 2006;19:779–87. doi: 10.1021/tx050326r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu B, Dietz BM, Dunlap T, Kastrati I, Lantvit DD, Overk CR, et al. Structural modulation of reactivity/activity in design of improved benzothiophene selective estrogen receptor modulators: induction of chemopreventive mechanisms. Mol Cancer Ther.;2007;6:2418–28. doi: 10.1158/1535-7163.MCT-07-0268. [DOI] [PubMed] [Google Scholar]
- 48.Okoh V, Deoraj A, Roy D. Estrogen-induced reactive oxygen species-mediated signalings contribute to breast cancer. Biochim Biophys Acta. 2011;1815:115–33. doi: 10.1016/j.bbcan.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 49.Loft S, Poulsen HE. Cancer risk and oxidative DNA damage in man. J Mol Med (Berl) 1996;74:297–312. doi: 10.1007/BF00207507. [DOI] [PubMed] [Google Scholar]
- 50.Chen Z-H, Na H-K, Hurh Y-J, Surh Y-J. 4-Hydroxyestradiol induces oxidative stress and apoptosis in human mammary epithelial cells: possible protection by NF-[kappa]B and ERK/MAPK. Toxicol Appl Pharmacol. 2005;208:46–56. doi: 10.1016/j.taap.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 51.Markides CS, Roy D, Liehr JG. Concentration dependence of prooxidant and antioxidant properties of catecholestrogens. Arch Biochem Biophys. 1998;360:105–12. doi: 10.1006/abbi.1998.0934. [DOI] [PubMed] [Google Scholar]
- 52.Park S-A, Na H-K, Kim E-H, Cha Y-N, Surh Y-J. 4-Hydroxyestradiol Induces Anchorage-Independent Growth of Human Mammary Epithelial Cells via Activation of kB Kinase: Potential Role of Reactive Oxygen Species. Cancer Res. 2009;69:2416–24. doi: 10.1158/0008-5472.CAN-08-2177. [DOI] [PubMed] [Google Scholar]
- 53.Zahid M, Saeed M, Beseler C, Rogan EG, Cavalieri EL. Resveratrol and N-acetylcysteine block the cancer-initiating step in MCF-10F cells. Free Radic Biol Med. 2011;50:78–85. doi: 10.1016/j.freeradbiomed.2010.10.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen ZH, Hurh YJ, Na HK, Kim JH, Chun YJ, Kim DH, et al. Resveratrol inhibits TCDD-induced expression of CYP1A1 and CYP1B1 and catechol estrogen-mediated oxidative DNA damage in cultured human mammary epithelial cells. Carcinogenesis. 2004;25:2005–13. doi: 10.1093/carcin/bgh183. [DOI] [PubMed] [Google Scholar]
- 55.Fu J, Fang H, Paulsen M, Ljungman M, Kocarek TA, Runge-Morris M. Regulation of estrogen sulfotransferase expression by confluence of MCF10A breast epithelial cells: role of the aryl hydrocarbon receptor. J Pharmacol Exp Ther. 2011;339:597–606. doi: 10.1124/jpet.111.185173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shatalova EG, Walther SE, Favorova OO, Rebbeck TR, Blanchard RL. Genetic polymorphisms in human SULT1A1 and UGT1A1 genes associate with breast tumor characteristics: a case-series study. Breast Cancer Res. 2005;7:R909–21. doi: 10.1186/bcr1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Steiner C, Peters WH, Gallagher EP, Magee P, Rowland I, Pool-Zobel BL. Genistein protects human mammary epithelial cells from benzo(a)pyrene-7,8-dihydrodiol-9,10-epoxide and 4-hydroxy-2-nonenal genotoxicity by modulating the glutathione/glutathione S-transferase system. Carcinogenesis. 2007;28:738–48. doi: 10.1093/carcin/bgl180. [DOI] [PubMed] [Google Scholar]
- 58.Rahal OM, Simmen RC. PTEN and p53 cross-regulation induced by soy isoflavone genistein promotes mammary epithelial cell cycle arrest and lobuloalveolar differentiation. Carcinogenesis. 2010;31:1491–500. doi: 10.1093/carcin/bgq123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Prossnitz ER, Barton M. The G-protein-coupled estrogen receptor GPER in health and disease. Nat Rev Endocrinol. 2011;7:715–26. doi: 10.1038/nrendo.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bolton JL, Yu L, Thatcher GRJ. Quinoids formed from estrogens and antiestrogens. Methods Enzymol. 2004;378:110–23. doi: 10.1016/S0076-6879(04)78006-4. [DOI] [PubMed] [Google Scholar]
- 61.Dowers TS, Qin ZH, Thatcher GRJ, Bolton JL. Bioactivation of selective estrogen receptor modulators (SERMs). Chem Res Toxicol. 2006;19:1125–37. doi: 10.1021/tx060126v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu L, Liu H, Li W, Zhang F, Luckie C, van Breemen RB, et al. Oxidation of raloxifene to quinoids: potential toxic pathways via a diquinone methide and o-quinones. Chem Res Toxicol. 2004;17:879–88. doi: 10.1021/tx0342722. [DOI] [PubMed] [Google Scholar]
- 63.Liu H, Liu J, van Breemen RB, Thatcher GRJ, Bolton JL. Bioactivation of the selective estrogen receptor modulator desmethylated arzoxifene to quinoids: 4′-fluoro substitution prevents quinoid formation. Chem Res Toxicol. 2005;18:162–73. doi: 10.1021/tx049776u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.