Fig. 1.
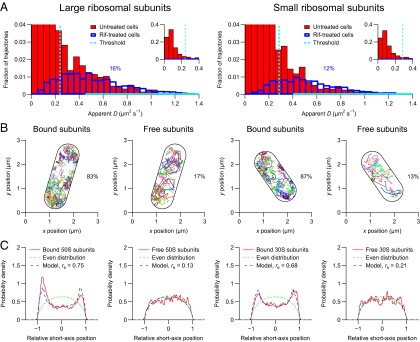
Tracking of individual ribosomal subunits in living E. coli cells. The cells were imaged at 50 Hz for 5 min on agarose pads with a laser excitation exposure time of 5 ms. The geometries of the imaged cells were determined from the positions of the individual ribosomal subunits. The lengths of the imaged cells were determined to be between 1.8 and 2.9 µm. Similar results are obtained if the geometries of the imaged cells are determined from out-of-focus bright-field images (SI Appendix, Fig. S9). (A) Distributions of apparent diffusion coefficients of individual large (Left) and small (Right) ribosomal subunits. The distributions in the untreated cells (red bars) are fitted with the corresponding distributions in rifampicin-treated cells (blue outlines) in the regions indicated by solid cyan lines. Each distribution corresponds to >1,000 trajectories from eight cells. (B) Trajectories of individual ribosomal subunits. Trajectories of free and mRNA-bound subunits are plotted separately. (C) Distributions of relative short-axis positions of bound and free ribosomal subunits in the cylindrical parts of eight cells (solid red curves) fitted with a model for nucleoid-excluded particles (dashed blue curves). Each distribution corresponds to >1,000 positions. The relative exclusion radius (re) is 0 for an evenly distributed particle (dashed green curves) and 1 for a membrane-bound particle.
