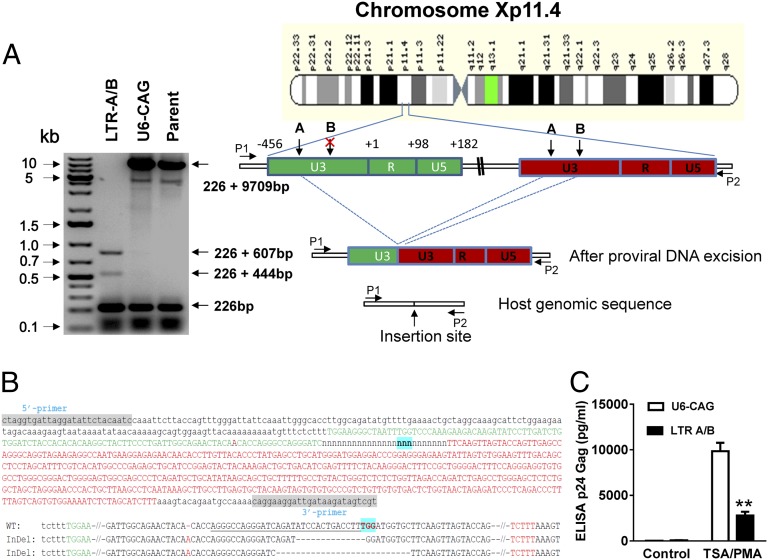
Fig. 2.
Cas9/LTR-gRNA efficiently eradicates latent HIV-1 virus from U1 monocytic cells. (A) (Right) Diagram showing excision of HIV-1 entire genome in chromosome Xp11.4. HIV-1 integration sites were identified using a Genome-Walker link PCR kit. (Left) Analysis of PCR amplicon lengths using a primer pair (P1/P2) targeting chromosome X integration site-flanking sequence reveals elimination of the entire HIV-1 genome (9,709 bp), leaving two fragments (833 and 670 bp). (B) (Upper) TA cloning and sequencing of the LTR fragment (833 bp) showing the host genomic sequence (small letters, 226 bp) and the partial sequences (634 − 27 = 607 bp) of 5′ LTR (green) and 3′ LTR (red) with a 27-bp deletion around the LTR A targeting site (underlined). (Lower) Two indel alleles identified from 15 sequenced clonal amplicons. The 670-bp fragment consists of a host sequence (226 bp) and the remaining LTR sequence (634-190 = 444 bp) after 190-bp excision by simultaneous cutting at LTR-A and -B target sites. The underlined and green-highlighted sequences indicate the gRNA LTR-A target site and PAM. (C) Functional analysis of LTR-A/B-induced eradication of HIV-1 genome, showing substantial blockade of p24 virion release induced by TSA/phorbol myristate acetate (PMA) treatment. U1 cells were transfected with pX260-LTRs A, B, or A/B. After 2-wk puromycin selection, cells were treated with TSA (250 nM)/PMA for 2 d before p24 Gag ELISA was performed.