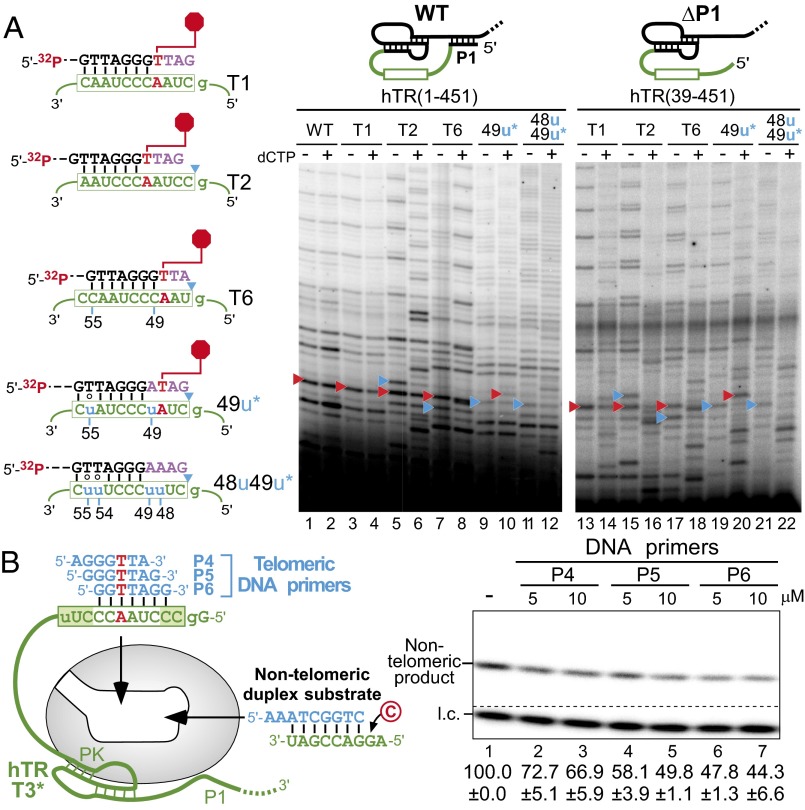
Fig. 4.
Functional assays of hTR template variants for sequence-defined pausing and binding affinity of the DNA/RNA duplex to the active site. (A, Left) Sequences of the DNA primer and the hTR template variants T1, T2, T6, 49u, and 48u49u. The sequence-defined pausing site (red hexagon) and the P1-defined boundary (blue triangles) are denoted. The asterisk (*) denotes that the 49u and 48u49u mutants contain additional mutations (55u and 54u55u, respectively) to permit processive repeat addition. (A, Right) Telomerase reconstituted in vivo with hTR template variants, T1, T2, T6, 49u, and 48u49u, were assayed with the 18-mer 32P end-labeled DNA primer (TTAGGG)3 in the presence (+) or absence (−) of 0.5 mM dCTP. The sequence-defined (red triangles) and P1-defined (blue triangles) pause sites are denoted. The secondary structure of full-length hTR (1–451) with helix P1 and truncated hTR (39–451) without helix P1 are shown above the gel. (B) Competitive inhibition assay of DNA primer competitors against a nontelomeric duplex substrate. Telomerase was reconstituted in vivo with the hTR template permuted variant T3* that lacks helix P1, the linker poly(U) tract, and contains a 56u mutation. The T3* telomerase was preannealed with excess P4, P5, or P6 telomeric DNA primers at 5 and 10 µM as competitors when assayed against 10 µM nontelomeric substrate with [α-32P]dCTP. A 32P end-labeled 7-mer oligonucleotide was included as a loading control (l.c.). Quantitation of the relative activity in the presence of competitor is displayed below the gel with the SE (±) derived from two independent experiments.