Figure 2.
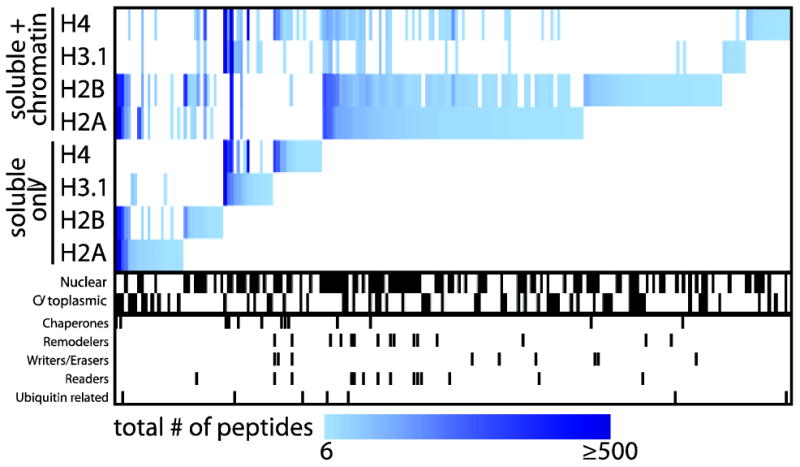
The use of DNA shearing in AP sample preparation enhances the identification by mass spectrometry of chromatin-bound interactors for core histone proteins. A heat map of the interaction partners identified with each histone bait using the “standard” or chromatin optimized AP approach (Avg SAINT ≥ 0.8) was generated using MeV 4.81. The pixel color corresponds to the total number of peptides (spectral counts) detected for a given prey across all replicates. Interactor localization and functions, as defined by GO annotation, is highlighted below the heat map.
