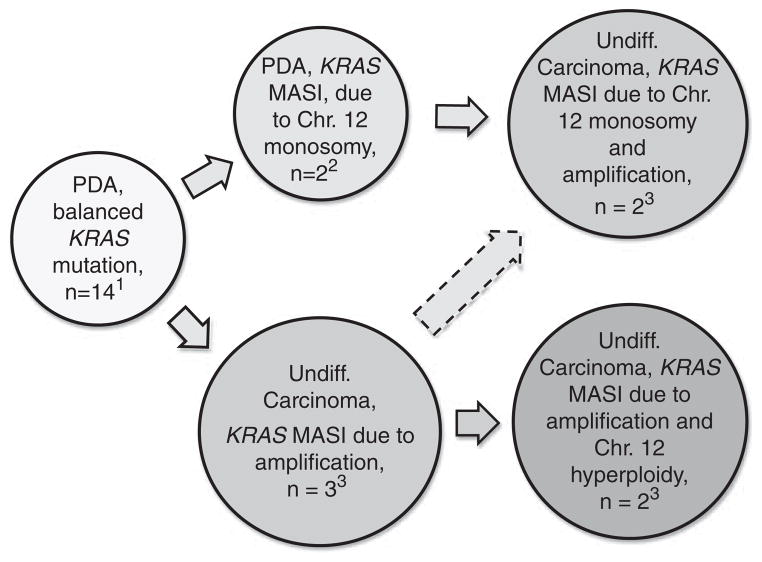
Figure 5.
KRAS copy number alterations and likely KRAS allelic imbalance associated with pancreatic ductal adenocarcinoma progression to undifferentiated pancreatic carcinoma. The scheme is based on previously published referenced work and a subset of cases presented in this report. Numbers refer to our cases for which both KRAS FISH and sequencing electropherograms were available. Note 1: The largest subset of pancreatic ductal adenocarcinoma (n=14) is characterized by normal results of KRAS FISH and no KRAS MASI. In these cases, the wild-type and mutated KRAS alleles are likely to be balanced and the fact that wild-type allele peak is higher than the mutant allele peak on sequencing electropherogram is best explained by the wild-type allele derived from stromal tissue and inflammatory cells. Note 2: Based on sequencing electropherograms, a subset of pancreatic ductal adenocarcinoma is characterized by KRAS MASI. KRAS FISH was performed on 5 of 6 such cases. Three cases showed normal KRAS FISH and KRAS MASI may have arisen through chromosome 12 uniparental disomy. In two cases, KRAS FISH revealed chromosome 12 monosomy in 47% and 78% of cells. As wild-type allele was predominant on sequencing electropherogram, the lost copy of chromosome 12 likely harbored the wild-type allele. Note 3: Sixteen of 25 undifferentiated carcinomas showed KRAS MASI. KRAS FISH was performed on 15 of 16 cases with KRAS MASI and amplification alone was identified in three cases. Two cases showed amplification associated with chromosome 12 monosomy, while the other two cases showed amplification associated with chromosome 12 hyperploidy.