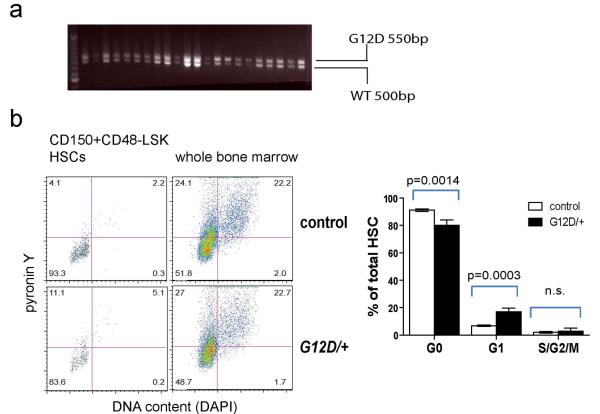
Extended data Figure 1.
a) The NrasG12D allele was recombined in all HSCs after 3 doses (every other day) of pIpC. Two weeks after the last dose of pIpC was administered to Mx1-cre; NrasG12D/+ mice, the mice were sacrificed and individual CD150+CD48−LSK HSCs were sorted into methylcellulose cultures in 96 well plates. The cells were cultured for 14 days then DNA was extracted from individual colonies and genotyped by PCR. The size of the recombined NrasG12D allele (G12D) was 550bp and the Nras+ allele (WT) was 500bp. Nras recombination was observed in 22 of 22 HSC colonies examined. Blot is representative of three independent experiments. b) Cell cycle analysis of HSCs by pyronin Y and DAPI staining. CD150+CD48−LSK HSCs were sorted from Mx1-cre; NrasG12D/+ mice and littermate controls into 100% ethanol and stained with pyronin Y and DAPI to identify cells in G0 (left lower quadrant), G1 (left upper quadrant) and S/G2/M (right upper and lower quadrants). Data represent mean±s.d.. Statistical analysis was performed with a two-way ANOVA (P<0.01, n=4) followed by pairwise posthoc t-tests.