Figure 4. Comparison of the HIV-1 drug-resistant mutations identified by standard population (Sanger) and deep sequencing.
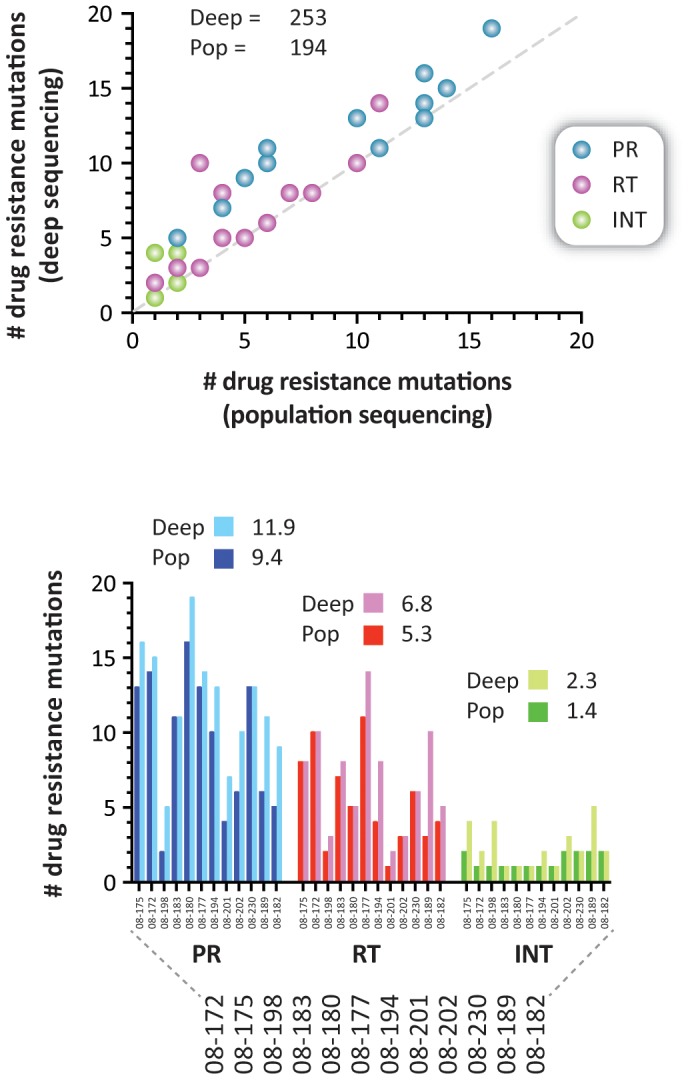
Plasma samples from the 12 treatment-experienced HIV-infected individuals participating in the GS-US-183-0105 study of elvitegravir were analyzed as described in Materials and Methods. The top plot compares the number of drug resistance mutations detected by Sanger and deep sequencing in each patient. The total numbers of drug resistance mutations identified by each sequencing method are indicated. The mean difference in the numbers of drug resistance mutations detected by population and deep sequencing in the protease (PR), reverse transcriptase (RT), and integrase (INT) regions is indicated in the bottom graph. Deep, deep sequencing; Pop, population sequencing.
