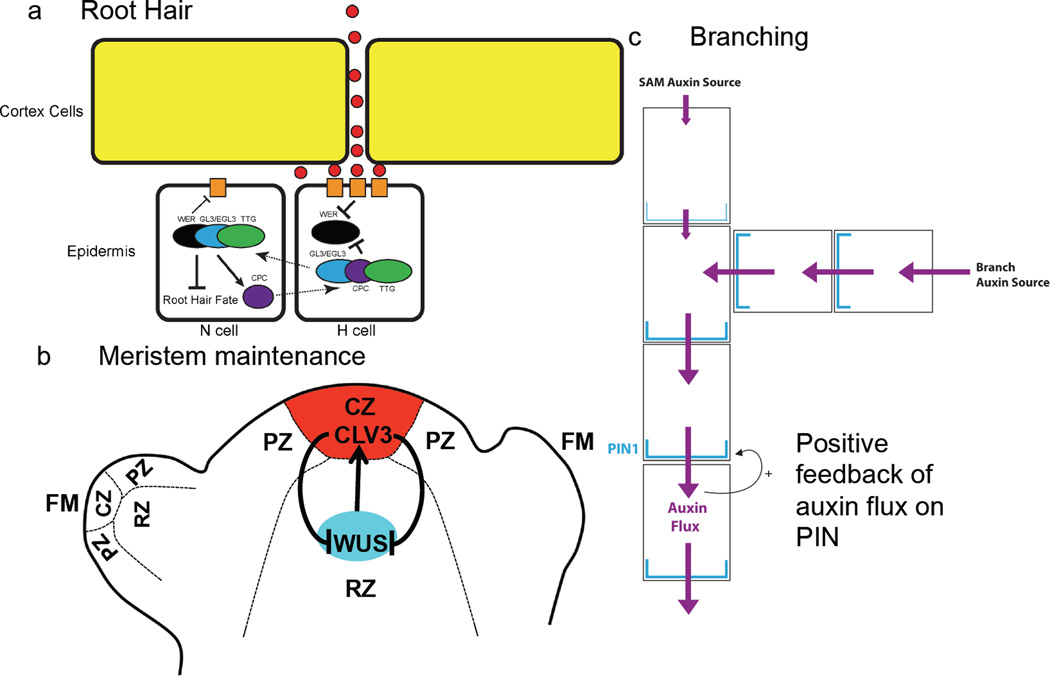
Figure 1. Gene regulatory networks?
aThe patterning of hair cell (H) and non-hair cells (N) in the root involves both spatial signaling through the SCRAMBLED receptor and activation or repression of transcriptional complexes that are influenced by the movement of proteins from one cell to another. H cell development is initiated by the combined effect of WER repression by activation of SCRAMBLED signaling by an unknown signal and CAPRICE (CPC) activity.
bThe SAM can be divided into the central zone (CZ), the peripheral zone (PZ) and the rib meristem (RM)62, 63. The CZ, comprises of a pool of pluripotent stem cells. The PZ is seated on the flanks of the SAM where new lateral organs are initiated. WUSCHEL (WUS) is expressed exclusively in the organizing center (RM) (blue) and is required to produce an unknown signal to specify the stem cell identity in the overlying cell layers. Stem cells in the CZ (red) secrete the CLAVATA3 (CLV3) small polypeptide, which activates a signalling cascade that limits the WUS transcription in the RM64. Thus, the negative feedback regulatory circuitry composed of WUS and CLV3 forms a self-correcting mechanism to maintain the stem cell homeostasis in the SAM.
cThe branching pattern of the plant can be understood by modelling the competition between auxin sources at the tip of each branch to transport auxin through regions of the plant stem (represented as large boxes). A model in which the flux of auxin (purple) positively feeds back on the amount of PIN1 auxin efflux transporter (blue) automatically establishes the competition between branches and reproduces the branching pattern of both wild type and mutant plants.