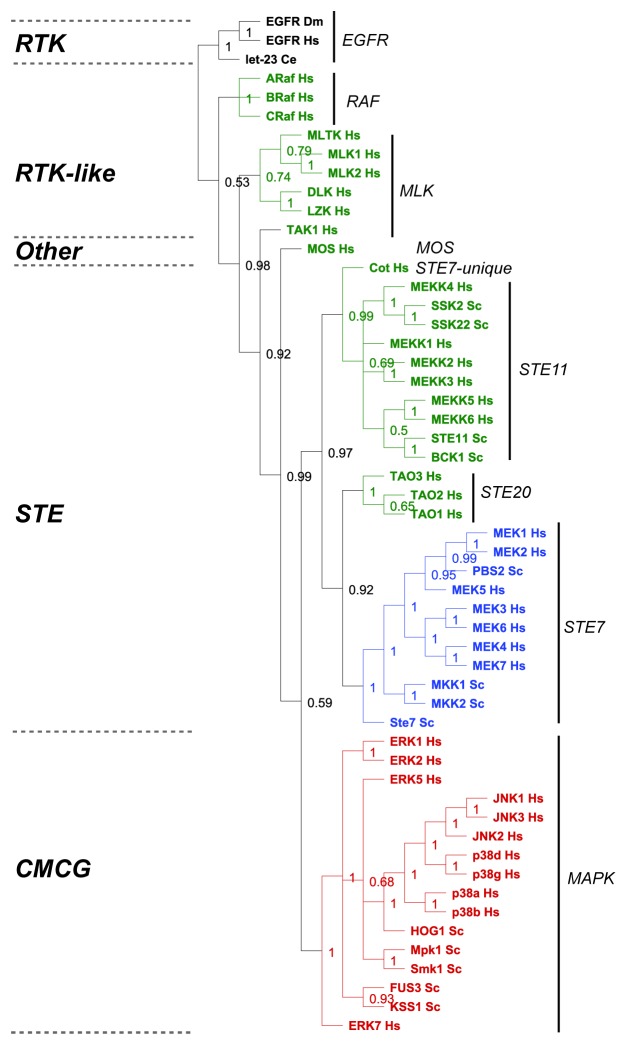
Figure 6. Evolutionary analysis of MAP kinase modules from yeast and man. We restricted our evolutionary analyses to kinases that have been experimentally verified to belong to MAP Kinase modules in humans (Hs) and baker's yeast Saccharomyces cerevisiae (Sc) (reviewed in refs. 5, 6, 27, and 28). We conducted a Maximum Likelihood evolutionary analysis on the aligned sequences using Mr Bayes,70 using the standard options available in the Geneious software plugin.71 The Epithelial Growth Factor Receptor (EGFR) from human, fruit fly (Drosophila melanogaster DM) and worm (let-23 Caenorhabditis elegans Ce) were used as out-groups to root the tree (shown in black). The M3K tier of the module is shown in green, the M2K tier shown in blue, and the M1K tier shown in red. The kinase family groups are indicated by lines and named on the right, the kinase groups are shown on the left.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.