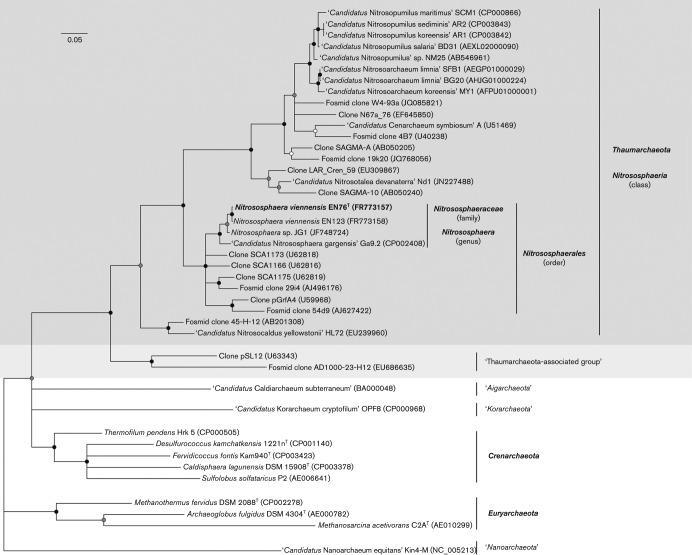
Fig. 5.
Maximum-likelihood 16S rRNA gene phylogeny of the Thaumarchaeota and representative strains of the Crenarchaeota, Euryarchaeota and other proposed archaeal phyla. The tree depicts Nitrososphaera viennensis EN76T (bold), the marine pure culture ‘Candidatus Nitrosopumilus maritimus’ SCM1, organisms from laboratory or natural enrichment cultures (labelled Candidatus) and a selection of environmental sequences representing major uncultured lineages. Proposed phyla and uncharacterized archaeal lineages are placed in quotes. Phylogeny reconstruction was based on 1202-bp 16S rRNA gene fragments and calculated with RaxML VI-HPC using the GTR+I+G model. Bootstrap support values (1000 replicates) are indicated by circles: filled, ≥90 %; shaded, ≥80 % but <90 %; open, ≥70 % but <80 %. Some branching points are not well supported in the displayed tree, such as the lineages of ‘Candidatus Caldiarchaeum’ and ‘Candidatus Korarchaeum’. The former was affiliated rather with Thaumarchaeota in more comprehensive phylogenetic calculations (see e.g. Eme et al., 2013).