Figure 3.
In Silico Protein Modeling
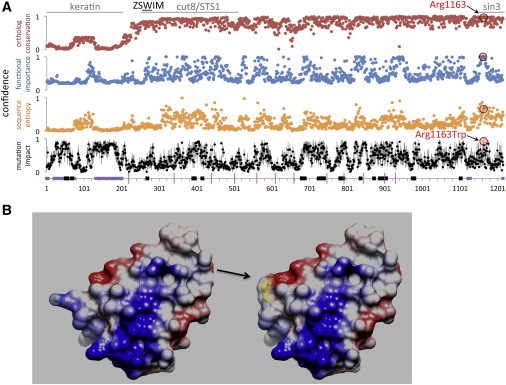
(A) One-dimensional analyses of the predicted functional relevance of protein domains in ZSWIM6. Arginine 1163 occurs in a highly conserved domain from 1148–1215 with similarity to Sin3. Substitution to tryptophan is predicted to have severe effects on protein function. Regions with detectable similarity to non-ZSWIM proteins are denoted at the top. One-dimensional analyses are plotted from the ZSWIM6 N terminus (residue 1, left) to C terminus (1215, right). Plots are shown for percent identity among 97 orthologous proteins (red), predicted functional importance (blue, measured by MFS), relative sequence entropy (orange, measured by HMMRE) and predicted impact of substitution (measured by HUSCY; the mean and standard deviation for all possible single-nucleotide variations are shown in black and gray). At bottom, predicted disordered regions (as measured by DISpro) are shown in purple, domain boundaries (determined by DOMpro) in black, and exon borders as pink lines. Scores for p.Arg1163 and p.Arg1163Trp are highlighted in red and black circles.
(B) Protein modeling of amino acid residues 1148–1215 of the wild-type (left) and mutant (right) shows that the p.Arg1163Trp substitution (arrow) dramatically alters the structure and hydrophobicity of the protein.
