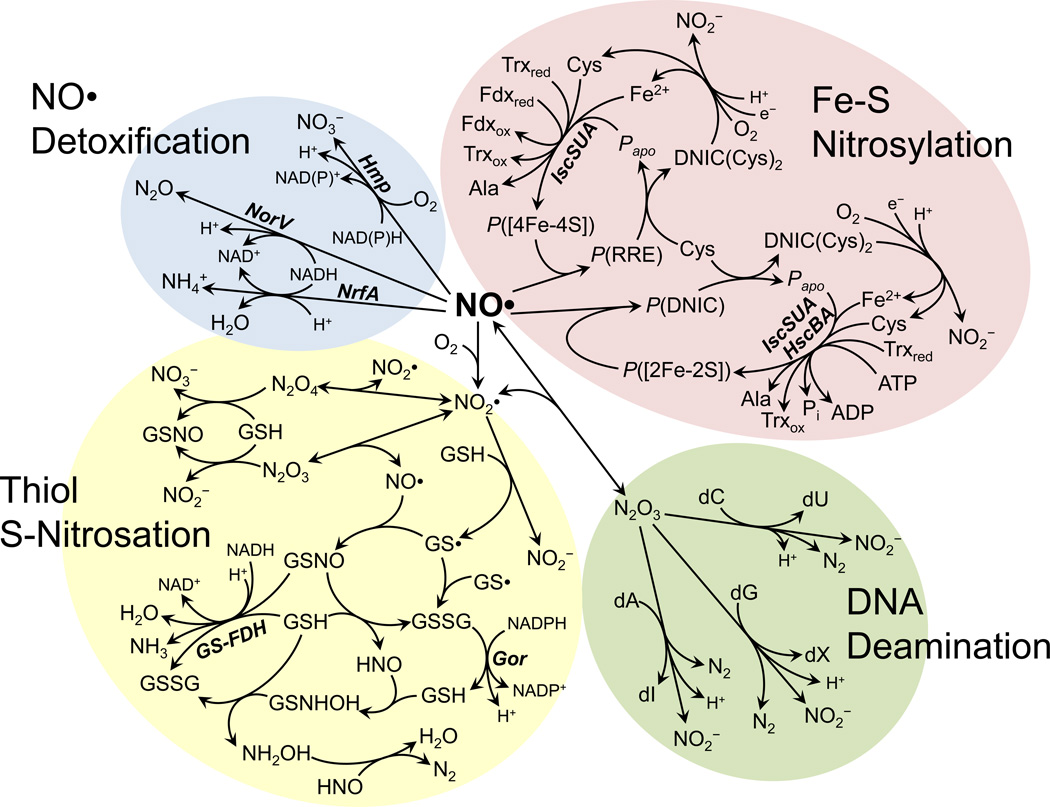
Figure 1.
Biochemical reaction network of NO• in E. coli. The diagram illustrates several key pathways involved in NO• metabolism. Reactions are grouped into categories of NO• detoxification, Fe-S nitrosylation and repair, DNA deamination, and thiol S-nitrosation and denitrosation. Enzymes catalyzing a reaction are shown in bold italics next to the reaction arrow. Enzyme abbreviations: NorV, NO• reductase; Hmp, NO• dioxygenase; NrfA, formate-dependent nitrite reductase; GS-FDH, glutathione-dependent formaldehyde dehydrogenase; Gor, glutathione reductase; IscSUA, Fe-S cluster assembly system; HscBA, Fe-S assembly chaperone. Metabolite abbreviations: GSH, glutathione; GS•, glutathionyl radical; GSNO, S-nitrosoglutathione; GSSG, glutathione disulfide; dA, deoxyadenosine; dG, deoxyguanosine; dC, deoxycytidine; dI, deoxyinosine; dX, deoxyxanthosine; dU, deoxyuridine; P([2Fe-2S]), protein-bound [2Fe-2S] cluster; P([4Fe-4S]), protein-bound [4Fe-4S] cluster; P(DNIC), protein-bound dinitrosyl-iron complex; P(RRE), protein-bound Roussins’ red ester; DNIC(Cys)2, L-cysteine-bound dinitrosyl-iron complex; Papo, apo-protein (lacking Fe-S cluster); Trxred, reduced thioredoxin; Trxox, oxidized thioredoxin; Fdxred, reduced ferredoxin; Fdxox, oxidized ferredoxin.