Abstract
Populus (Salicaceae) is one of the most economically and ecologically important genera of forest trees. The complex reticulate evolution and lack of highly variable orthologous single-copy DNA markers have posed difficulties in resolving the phylogeny of this genus. Based on a large data set of nuclear and plastid DNA sequences, we reconstructed robust phylogeny of Populus using parsimony, maximum likelihood and Bayesian inference methods. The resulting phylogenetic trees showed better resolution at both inter- and intra-sectional level than previous studies. The results revealed that (1) the plastid-based phylogenetic tree resulted in two main clades, suggesting an early divergence of the maternal progenitors of Populus; (2) three advanced sections (Populus, Aigeiros and Tacamahaca) are of hybrid origin; (3) species of the section Tacamahaca could be divided into two major groups based on plastid and nuclear DNA data, suggesting a polyphyletic nature of the section; and (4) many species proved to be of hybrid origin based on the incongruence between plastid and nuclear DNA trees. Reticulate evolution may have played a significant role in the evolution history of Populus by facilitating rapid adaptive radiations into different environments.
Introduction
The genus Populus, distributed throughout the northern hemisphere from subtropical to boreal forests [1] and one of the most economically and ecologically important genera of forest trees [2], is well known for its rapid growth, profuse vegetative propagation, environmental stress tolerance and the numerous uses of its wood [3]. Furthermore, the genus has become an excellent research model due to its small genome size and the completion of the genome sequence of P. trichocarpa [4]. A clear understanding of the evolutionary relationships of Populus species will provide an important foundation for biological studies and genetic breeding programs.
The relationships among sections as well as relationships within each section remain controversial and/or poorly resolved because of the extensive interspecific hybridization and high degree of morphological variation among species [3], [5]. The combination of these two features results in a major disagreement in the number of species and their delimitation [5], [6]. Eckenwalder [5] recognized 29 species of Populus grouped into six sections (Abaso, Aigeiros, Leucoides, Populus, Tacamahaca, Turanga) based on morphological similarity and crossability. However, more than 60 species (plus a number of hybrids, varieties and forms) are described in the Flora of China [6]. The morphology-based phylogenetic tree of Populus demonstrated that section Abaso and Turanga were sister groups to the other four sections followed by section Leucoides, but the relationship among the other three sections remain unresolved [5]. Phylogenetic analysis based on 5.8S RNA and ITS sequences suggested that section Populus was sister to Leucoides, Tacamahaca and Aigeiros [7]. On the other hand, a phylogenetic tree based on plastid RFLP data showed an opposite trend with section Populus as an advanced clade occupying the terminal position [8]. Cervera [9] proposed that P. lasiocarpa and P. violascens of section Leucoides should be classified into section Tacamahaca. Furthermore, the systematic placement of species in section Aigeiros is inconsistent [8], [10]. Cervera [9] proposed that P. nigra should be classified into a new section or as a subsection of section Tacamahaca. Most interspecific relationships within each section, in particular species in sections Populus, Tacamahaca and Aigeiros, are poorly resolved.
In addition to the lack of highly variable orthologous single-copy DNA markers, the complex reticulate evolution in Populus poses difficulty in resolving the phylogeny of Populus [8], [10], [11]. Species of Populus show extensive hybridization within sections as well as between closely-related sections. The species of section Aigeiros, Tacamahaca and Leucoides can intercross freely [12], [13]. The evidence for hybridization in Populus has been documented based on molecular markers, for example, in the phylogenetic study based on maternally inherited plastid and biparentally inherited nuclear DNA sequences, P. nigra showed different affinity to sections Populus and Tacamahaca, which suggested a possible hybrid origin for P. nigra [8], [10]. P. tomentosa has also been suspected to be a hybrid for a long time, but its exact parents remain unknown. The inconsistent systematic position of section Populus mentioned above may also be an indication of ancient hybridization in Populus.
The phylogenetic topology derived from maternally inherited plastid [14] data sets represents the maternal genealogy while nuclear DNA phylogeny mirrors biparental evolutionary history. Comparative analysis of DNA sequences from the nuclear and the plastid offers an effective way to parse out reticulate evolutionary events [8], [15]. The completion of the whole genome of P. trichocarpa [4] provides a means to find highly variable single-copy nuclear DNA sequences to assess the evolutionary relationships of Populus. Here, we utilized 24 single-copy nuclear DNA sequences and 12 plastid fragments to reconstruct the phylogeny of Populus with an emphasis on the reticulate evolution in the genus.
Materials and Methods
Ethics Statement
No special permits were required for this study and this study did not involve endangered or protected species.
Species sampling, DNA extraction, PCR amplification and sequencing
The sampling was based on the classification proposed by Eckenwalder [5] and the Flora of China [6]. During the fieldwork from 2005 to 2013, we sampled twenty-six species representing 5 sections of Populus and 4 species of Salix as outgroups for the phylogeny reconstruction. The fresh leaves were dried and stored in silica gel. Information about the sampled species is given in (File S1 in Supporting Information S1).
Using a high throughput comparative genomic approach, Duarte et al. [16] identified 959 single-copy genes that were shared among P. trichocarpa, Arabidopsis thaliana, Vitis vinifera and Oryza sativa from which fifteen sequences were selected and characterized in our previous paper (Du et al., in press). The remaining nine pairs of primers were developed following the methods as described in Du et al. (in press). Twelve pairs of primers were selected from previous studies [17]–[21] and used for plastid amplification. All the primers used for amplifying and sequencing are listed in (File S2 in Supporting Information S1).
Total genomic DNA was isolated from silica-gel-dried leaves using a modified method [22]. Polymerase chain reaction (PCR) was performed in a total volume of 30 µL containing 5–50 ng of genomic DNA, 3 µL 10×PCR Buffer, 2.0 mM MgCl2, 0.8 µM of each dNTP, 2.4 µM of each primer and 0.15 U Ex Taq DNA polymerase (TaKaRa, Shiga, Japan). Amplification was carried out in a temperature gradient 96 U thermocycler (Applied Biosystems, Forster City, CA, USA), using following thermal cycling profiles: 4 min at 95°C followed by 30 cycles of 30 s at 94°C, 30 s at 52°C to 60°C (depending on the optimal annealing temperature of specific primers), 90 s at 72°C and a final extension at 72°C for 10 min. After purifying using a DNA Purification kit (Amersham Pharmacia Biotech, Piscataway, USA), the PCR product was sequenced using an ABI 3730 DNA analyzer (Applied Biosystems, Foster City, California, USA) with the same primers used for amplification. For the samples where direct sequencing failed, the purified PCR products were cloned into pGEM -T easy Vector System II (Promega, Madison, WI, USA). For each sample, 6–12 positive clones were randomly picked and sequenced in both directions using standard T7 and SP6 primers.
Data analysis
The assembled contigs of each individual were aligned using CLUSTAL X [23] and refined manually in BioEdit [24]. For all the loci, regions with more than 5 mononucleotide or microsatellite repeats were excluded because of the uncertainty of homology which could be exacerbated by potential inaccuracies of enzymatic process during PCR and sequencing [25], [26]. All indels were excluded in the following phylogenetic analyses after coded as binary characters according to the simple indel coding method [27] using FastGap 1.2 [28].Phylogenetic relationships among species were reconstructed using parsimony, maximum likelihood (ML) and Bayesian methods. The parsimony analysis was conducted in PAUP* 4.0b10* [29], with all characters equally weighted and treated as unordered. Heuristic search was performed with MULPARS option, tree-bisection-reconnection (TBR) branch swapping, RANDOM stepwise addition with 1000 replicates and the number of trees held in RAM was set to be 100000. Bootstrap analysis was conducted to assess topological robustness with 1000 replicates using simple taxon addition [30]. An appropriate nucleotide substitution model for each sequence was determined using jModeltest [31]. The models were chosen according to the Akaike information criterion (AIC) and used for subsequent ML and Bayesian analysis. ML analysis was conducted in PAUP* 4.0b10* [29] with random taxon addition of 1000 replicates, TBR branch swapping, MULPARS option, 100000 trees held in RAM and 100 replication of bootstrap analysis. Bayesian inference was performed with MrBayes 3.2.1 [32]. Two independent runs of Metropolis-coupled MCMC were conducted simultaneously, with each run being one cold chain and three incrementally heated chains and all started randomly in the parameters space. All other parameters were set to default. 1,000,000 generations were run and trees were sampled once every 100 generations. The program Tracer v1.5 [33] was utilized to check for stationary. The first 25% of sampled trees were discarded as burn-in and the posterior probabilities were calculated from the remaining trees. All the phylogenetic trees were viewed in the program FigTree v 1.3.1 [34]. The homogeneity across nuclear DNA loci was tested using the Shimodaria-Hasegawa test [35] in CONSEL [36].
Results
Sequence Characteristics
We successfully obtained all 24 nuclear DNA sequences and 12 plastid fragments from all 30 species except for the locus DSH22 in S. raddeana and DSH11 in P. grandidentata, which failed to amplify and were treated as missing data in the subsequent phylogenetic analyses. After removing regions with mononucleotide repeats and microsatellite sequences, the aligned length of the nuclear DNA ranged from 222 bp to 1106 bp with a total length of 15732 bp, in which exon sequences consisted of 10184 bp (64.7%). As shown in (File S3 in Supporting Information S1), the number of variable sites ranged from 38 (locus DSH6) to 160 (locus DSH10) and that of informative sites ranged from 27 (locus DSH6) to 111 (DSH22). The aligned length of plastid fragments varied between 532 bp and 2620 bp with a total length of 14197 bp, the exon sequences of which only occupied 4592 bp (32.3%). The most appropriate models fitted each locus decided by jModeltest [31] was presented in (File S3 in Supporting Information S1).
Phylogenetic analysis of the plastid fragments
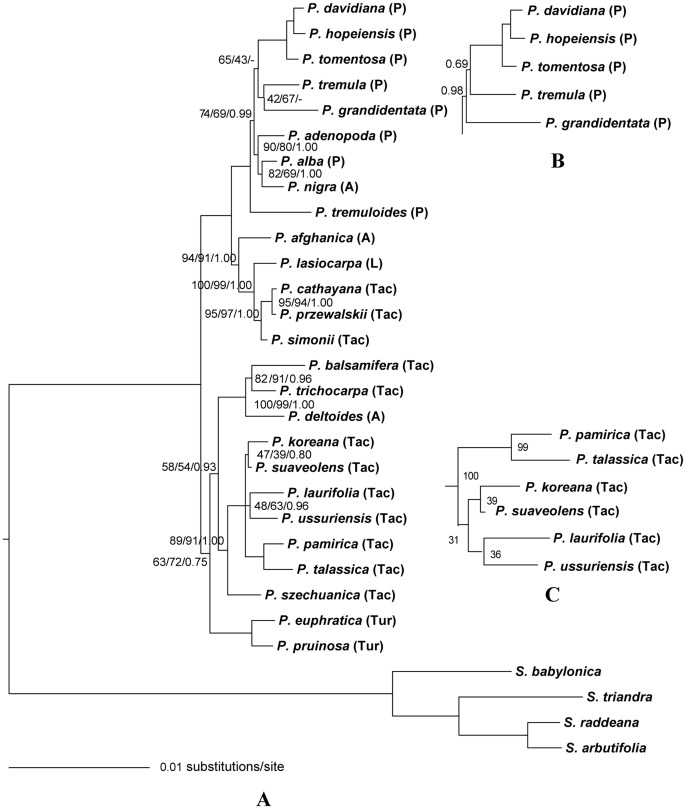
Phylogenetic relationships among species remained poorly resolved based on individual plastid sequences (File S5 in Supporting Information S1). Because all plastid gene sequences are effectively inherited as one locus, they were concatenated into a single contiguous sequence for the phylogenetic analysis. The best fitting evolutionary model for the combined plastid data set was TVM+G in ML and Bayesian analyses. 50% majority-rule consensus parsimony and ML trees were generated from 8 most parsimonious trees and 6 most likely trees based on the combined plastid data set. The phylogenetic trees generated by parsimony, ML and Bayesian methods were similar to each other, with only a few differences in bootstrap support (BS) or posterior probability (PP) values in some clades. For instance, the sister relationship between P. tremula and P. grandidentata, and between P. tomentosa and P. tremula/P. grandidentata in the parsimony and ML trees were not supported by Bayesian methods (Figure 1).
Figure 1. Phylogeny of Populus obtained from the combined 12 plastid fragments using parsimony method, (A).
Numbers next to nodes sequentially indicated ML/parsimony/BI support values. The branches without numbers indicate 100% statistical support. (B) The topology difference derived from Bayesian analysis while (C) is from ML analysis. A, Aigeiros; L, Leucoides; P, Populus; Tac, Tacamahaca; Tur, Turanga.
As shown in Figure 1, all Populus species formed a fully supported monophyletic group comprising two major clades. In the first clade, all species of section Populus and P. nigra of section Aigeiros formed a single group with P. tremuloides sister to other species. Within this group, P. davidiana, P. hopeiensis and P. tomentosa grouped together sister to P. tremula and P. grandidentata, meanwhile, P. alba, P. nigra and P. adenopoda showed close phylogenetic relationships to each other. Unexpectedly, P. cathayana, P. simonii and P. przewalskii of section Tacamahaca, P. lasiocarpa of section Leucoides and P. afghanica of section Aigeiros formed the other highly supported group. In the second clade, P. pruinosa and P. euphratica of section Turanga clustered together as sister taxa to other species. Three American poplars, P. balsamifera, P. trichocarpa and P. deltoides were more closely related to each other than to other Asiatic Tacamahaca species which further divided into two subclades with P. szechuanica sister to a group of species comprising P. koreana, P. suaveolens, P. laurifolia, P. ussuriensis, P. pamirica and P. talassica.
Phylogenetic analysis of the nuclear DNA
Phylogenetic relationships based on individual nuclear DNA loci were not fully resolved (File S6 in Supporting Information S1). The Shimodaria-Hasegawa test showed that there was no significant incongruence among most of the individual nuclear DNA loci (P>0.05), but slight incongruence was detected in some cases (File S4 in Supporting Information S1). After excluding these incongruent loci from the phylogeny reconstruction, topology of the phylogenetic tree remained the same with only some differences in BS or PP value. We, therefore, combined the 24 individual nuclear DNA sequences to a single data set to reconstruct the phylogeny of Populus and to make direct comparison with the cpDNA phylogeny.
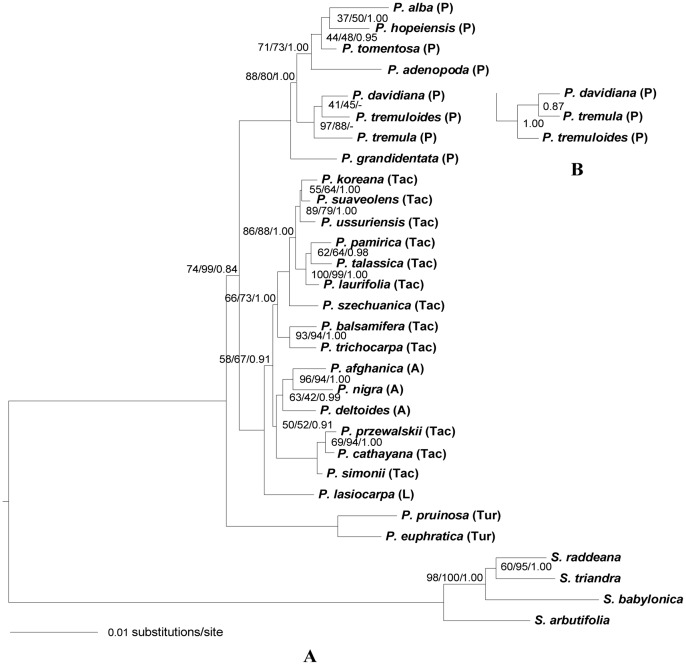
The best fitting evolutionary model for the combined nuclear DNA data set was GTR+R in ML and Bayesian analyses. The topologies of parsimony and ML trees are from 50% majority-rule consensus trees, which were generated from 4 most parsimonious trees and 6 most likely trees, respectively, based on combined nuclear DNA data set. Nuclear DNA phylogenetic trees generated from parsimony, ML and Bayesian methods showed similar topologies to each other with the only differences within section Populus of Bayesian tree (Figure 2). In the phylogenetic tree generated using nuclear data set (Figure 2), monophyly of Populus was strongly supported. Section Turanga was sister to other sections, followed by section Populus with high support value, in which a native North America poplar, P. grandidentata was sister to other species. The remaining species of section Populus subdivided into two groups: P. tomentosa, P. alba, P. hopeiensis and P. adenopoda clustered together with slightly lower resolution in the first group and three tremble aspens, P. davidiana, P. tremuloides and P. tremula showed close relationship to each other with high statistical support in the second. Section Leucoides (P. lasiocarpa) followed section Populus and was sister to sections Tacamahaca and Aigeiros. All the species of these two sections subdivided into two subclades. In the first subclade, P. cathayana, P. simonii and P. przewalskii clustered together sister to section Aigeiros, within which all the three species clustered as a single clade. The second subclade consisted of two sister groups: the two North America species (P. balsamifera/P. trichocarpa) and the other 7 species.
Figure 2. Phylogeny of Populus obtained from the combined 24 single-copy nuclear DNA sequences using ML method, (A).
Numbers next to nodes sequentially indicate ML/parsimony/BI support values. The branches without numbers indicate 100% statistical support. (B) The topology difference derived from Bayesian analysis. A, Aigeiros; L, Leucoides; P, Populus; Tac, Tacamahaca; Tur, Turanga.
Discussion
Phylogenetic relationships of Populus
With a large combined data set of 24 nuclear DNA sequences and 12 plastid fragments, we reconstructed maternal and biparental phylogenies of Populus (Figure 1 and 2). All species of Populus clustered together as one clade separated from outgroup species, which supported the results of previous studies [5], [10], [37], [38]. Species of each section grouped together in both nuclear and plastid DNA phylogenies with certain exceptions. Species of section Aigeiros clustered within different clades in plastid tree. P. simonii, P. cathayana and P. przewalskii of section Tacamahaca grouped with P. lasiocarpa separating from other balsam poplars in the plastid phylogeny.
There has been an argument about the oldest lineage of Populus for a long time. Phylogenetic analysis based on AFLP and ITS sequence proposed that section Populus was the oldest lineage in Populus [9]. However, fossil records and morphological phylogenetic analysis showed that section Turanga was sister to other sections [5]. Populus wilmottae, one of the most ancient fossil species of Populus [39], bears three-valved capsules, a feature considered primitive based on its predominance in the Violales, including many Flacourtiaceae, which is shared by section Turanga [5], [39]. Furthermore, the modern Turanga lineage is strikingly heteroblastic, with willow-like juvenile leaves strongly differentiating it from other species of Populus — thought to have developed early in the evolutionary history of this genus [5]. This suggested that section Turanga is an ancestral lineage as observed in phylogenetic trees with high BS or PP support value.
Intersectional relationships of Populus have been poorly resolved because of inadequate samples and/or insufficient resolution of molecular markers used in previous studies. Eckenwalder [5] reconstructed the phylogeny of Populus using 76 morphological characters. Section Abaso and Turanga were sister to other sections followed by section Leucoides and the relationship among the three advanced sections remained unresolved [5]. In an ITS sequence-based phylogeny of Populus, section Populus was monophyletic and species of sections Tacamahaca and Aigeiros mixed together in one clade with low statistical support [40].The relationship between the two sections did not resolve in trnT-trnF phylogeny [10]. In the phylogenetic tree reconstructed based on AFLP data, Cervera et al. [9] found that section Leucoides showed close relationships with sections Tacamahaca and Aigeiros. Moreover he proposed that P. ciliata, P. lasiocarpa and P. violascens of section Leucoides should be classified into section Tacamahaca [9]. ISSR-based phylogeny of Populus also revealed close intersectional relationship between section Tacamahaca and Aigeiros [37].
Based on the nuclear DNA phylogeny, section Populus and Leucoides were successive to Turanga, section Tacamahaca and Aigeiros occupied the terminal position. This phylogenetic order more or less followed previously published patterns [5]. The close affinity between section Tacamahaca and Aigeiros observed in the present study is in agreement with previous studies [5], [10], [32], [34]. For the first time, we found that species of Populus divided into two major clades in the plastid phylogeny, which reflected an early divergence of the maternal progenitors of Populus.
Intrasectional phylogenetic relationships
In previous studies, interspecific relationships within sections of Populus were rarely addressed or poorly resolved [9], [10]. Based on our data, intrasectional relationships among species were relatively better resolved. Because only one species of section Leucoides and two species of section Turanga were utilized in analysis, interspecific relationships of these two sections cannot be assessed. Furthermore, interspecific relationships of section Aigeiros is discussed below with an emphasis on hybrid origin.
Section Populus
The results from both nuclear and plastid DNA analyses suggest that section Populus is monophyletic. In the nuclear DNA tree, P. grandidentata was sister to other species which subdivided into two clades. The North American trembling aspen P. tremuloides showed close genetic affinity to Eurasian aspen P. tremula and P. davidiana with high BS or PP value. These three species are similar to each other with respect to morphological characters [6], [41]. Eckenwalder [5] even proposed that P. tremuloides, P. tremula and P. davidiana should be merged into a single species. The rationalization of this hypothesis and the accurate time of origin and differentiation among the three species require further analysis with a larger sample size. The remaining four species group in a single clade in keeping with their morphological similarity. Unlike the nuclear DNA tree, another North American aspen, P. tremuloides, separated from other species and formed a single clade in the plastid tree, which indicated that P. tremuloides was of an ancestral maternal origin. As in the nuclear DNA phylogeny, P. adenopoda and P. alba show closest affinity to each other whereas they are widely distributed in southern China and central Eurasia, respectively. It is of great interest to pay attention to allopatric speciation resulted from geographic isolation between these two species in future.
Section Tacamahaca
The relationships of species within section Tacamahaca are known to be the most complicated. In present study, P. cathayana, P. przewalskii and P. simonii clustered with section Leucoides in the plastid-based tree but sister to section Aigeiros in nuclear DNA tree as a single highly supported group. The classification of P. cathayana, P. przewalskii and P. simonii either in section Tacamahaca or in other sections requires further investigation based on comprehensive sample with morphological and molecular methods. The other balsam poplars formed two sister clades. Two North American balsam poplars P. trichocarpa and P. balsamifera formed a strongly supported clade sister to the Asian ones in the plastid and nuclear DNA phylogeny considering their morphological, ecogeographic similarity and recent divergence [42]. A similar situation was shown in Asiatic species of section Tacamahaca; however, their distribution is allopatric in China. We speculate that these species derived from vicariance and allopatric divergence from a once widely-distributed ancestral species of section Tacamahaca. The polyphyly of section Tacamahaca was clearly verified based on our plastid and nuclear DNA phylogeny [10], which was in agreement with morphology-based phylogenetic analysis [5].
Reticulate evolution in Populus
The most ancient undisputed fossil record of Populus dated to late Paleocene (about 58 Ma) is considered to be related to section Turanga because of the morphological similarity [39], [43]. Through combined fossil record and phylogenetic analyses we ascertain that section Turanga is more primitive in Populus. Species of the section Leucoides which inhabit permanent swamps first appeared in North America in late Eocene [5]. Combining fossil records and the phylogenetic tree based on 76 morphological characters, Eckenwalder [5] speculated that the temperate habitats of Populus were invaded by an ancestral member of section Leucoides, and following this, there was a rapid radiation (effectively simultaneous) driven by ancient hybridization events into the distinct habitats along with appearance of other advanced sections. The newly appeared sections can adapt to more extreme environments, for example, section Populus can tolerate aridity and coldness. The subsequent evolution within each section were partly influenced by hybridization [1].
Section Populus
Based on our plastid and nuclear DNA phylogeny, the ancestor of section Populus originated from the hybridization of two ancestral sections (section Turanga and Leucoides) with section Leucoides as the maternal parent. In the plastid tree, P. tremuloides was sister to other species in section Populus. In the nuclear DNA tree, P. tremuloides clustered with the Asiatic trembling aspen. According to leaf morphological variation and the fossil record, P. tremuloides was hypothesized to be a species of complex origin derived from multiple ancestral hybridization [5], [44]. It is inferred that hybridization occurred between ancestors of P. davidiana and P. tremula dispersed from Eurasia to North America as the paternal parent and P. tremuloides.
P. tomentosa, one of the cultivated species widely distributed in China, has attracted attention from taxonomists and geneticists [45]. Based on morphological traits and molecular evidence, P. tomentosa is considered as a complex hybrid species involving more than two species. Both RAPD and AFLP analysis suggests that P. tomentosa has closest affinity to P. adenopoda and was possibly a natural hybrid of P. alba and P. adenopoda [46]. The nuclear DNA phylogeny also revealed the close relationship between P. tomentosa and P. adenopoda. However, P. adenopoda and P. tomentosa clustered in two different clades within section Populus in the plastid phylogeny. In the hybridization event giving rise to P. tomentosa, the ancestor of P. davidiana and P. hopeiensis served as the maternal parent and P. adenopoda as the paternal role. Based on morphological similarity to P. hopeiensis and the closer genetic affinity, we infer that P. tomentosa may have been domesticated from P. hopeiensis, which shows sympatric distribution in China.
Section Tacamahaca
Twelve species of section Tacamahaca were used in this study. P. cathayana, P. przewalskii and P. simonii clustered with section Leucoides based on plastid phylogeny and sister to section Aigeiros in nuclear DNA tree, which indicated that their maternal lineage derived from section Leucoides followed by high gene flow with section Aigeiros. The remained nine species of section Tacamahaca showed a close affinity to section Turanga in the plastid tree and sister to section Leucoides in nuclear DNA tree, which suggested these species also derived from a hybridization event in which section Turanga and section Leucoides played the maternal and paternal roles, respectively.
Section Aigeiros
Three species of section Aigeiros were used in this study. They clustered together in the nuclear DNA tree, however, in the plastid tree P. nigra clustered with P. alba, P. deltoids grouped with P. balsamifera and P. trichocarpa while P. afghanica showed close affinity to P. lasiocarpa. This implied that all of the three species of section Aigeiros were hybrid origin with different maternal parent.
Despite large number of morphological, molecular and phylogenetic studies on the hybrid origin of P. nigra, the maternal parent of this species remains uncertain. Phylogenetic analysis based on plastid data clustered P. nigra within section Populus and the plastid of P. nigra show close affinity to either P. alba [8] or common ancestors of P. tremula and P. davidiana [10]. Our results confirm that the plastid of P. nigra is inherited from P. alba or a common ancestor shared with P. alba. A close relationship between P. deltoides and section Tacamahaca was observed previously [9], [10], [37], [47]. In the plastid tree, P. deltoides along with two native North American balsam poplars of section Tacamahaca form a monophyletic group with high BS or PP values. The similarity of floral morphology [48], overlapping distribution [49] and hybridization [50]–[52] of these three species in North America suggest that they may have diverged from a common maternal ancestor following a hybridization event giving rise to P. deltoides. A similar situation was also seen in P. afghanica, which shares a common maternal ancestor with P. lasiocarpa.
The taxonomic position of species in section Aigeiros is contentious. Rajora & Dancik [47] proposed a new section Nigrae, consisting only of P. nigra. Cervera et al. [9] pointed out that P. deltoides should be separated from the consectional P. nigra. Moreover, Eckenwalder [53] suggested that sections Tacamahaca and Aigeiros should be merged into a single section because of their close evolutionary relationships, which was supported by phylogenetic analysis based on ITS and plastid trnT-trnF sequences of Populus [10]. Nevertheless, considering the highly supported relationship in our nuclear DNA phylogeny and morphological similarity among the species of section Aigeiros, it is reasonable to retain this section which is a good model for further research about hybrid speciation in Populus.
In all, species of section Populus and Tacamahaca played a part in the origin of species in section Aigeiros, which suggests that section Aigeiros may have originated later than the former two sections. This is consistent with the paleontological results, and fossils of the section Aigeiros occur later in the sediments [5].
Conclusions
Based on nucleotide sequences of 24 single-copy nuclear genes and 12 plastid fragments, two robust phylogenies of Populus (Salicaceae) were reconstructed. The genus Populus was monophyletic in both phylogenetic trees and section Turanga was an ancestral lineage within the genus Populus. Comparative analyses of these two phylogenetic trees revealed reticulate evolutionary patterns in this genus. Three advanced sections (Populus, Aigeiros and Tacamahaca) were of hybrid origin. A detailed study involving more species (especially section Abaso) and genes are needed to further infer the origin, dispersal and hybridization in Populus.
Supporting Information
Combined supporting information file. File S1. The information of species (names, altitude, longitude, latitude, collector, collection number and place of voucher deposition). File S2. The information of primers. File S3. The information of nuclear and plastid DNA sequences. File S4. Results of the Shimodaira-Hasegawa test that are significant incongruence (P<0.05). File S5. Phylogenetic trees reconstructed based on plastid fragments using parsimony, most likelihood and Bayesian inference methods. Numbers next to nodes indicated bootstrap support value or posterior probabilities. File S6. Phylogenetic trees reconstructed based on nuclear DNA fragments using parsimony, most likelihood and Bayesian inference methods. Numbers next to nodes indicated bootstrap support value or posterior probabilities.
(RAR)
Acknowledgments
We thank Dr. Ge Song, Dr. Zhang Fumin, Dr. Zou Xinhui and Dr. Tang Liang in State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing, China, for their suggestion on data analysis. We also thank Li Yunxiao and Li Shanshan in Zhang's group for data analysis and experimental work. We appreciate two anonymous reviewers who gave valuable advice on this paper. Data archiving: Sequence data used in this study have been submitted to GenBank: accession numbers KF940053-KF941130.
Funding Statement
Financial support for this research was provided by Key Project of Research Institute of Forestry, Chinese Academy of Forestry (ZD200911), the Special Research Program for Public-welfare Forestry of China (201004035), corporation project on science and postgraduate education in Beijing and collaborative innovation plan of Jiangsu higher education. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.DiFazio SP, Slavov GT, Joshi CP (2011) Populus: a premier pioneer system for plant genomics. In: Joshi CP, editor. Genetics, genomics and breeding of poplar. Lebanon: Science Publishers, Inc. pp: 1–28. [Google Scholar]
- 2.Stettler RF Zsuffa L, Wu R (1996) The role of hybridization in the genetic manipulation of Populus. In: Stettler RF, Bradshaw HD, Heilman PE, Hinckler TM editors. Biology of Populus and its implications for management and conservation. Ottawa: Canadian Government Publishing. pp. 87–112. [Google Scholar]
- 3. Cronk Q (2005) Plant eco-devo: the potential of poplar as a model organism. New Phytologist 166: 39–48. [DOI] [PubMed] [Google Scholar]
- 4. Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, et al. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604. [DOI] [PubMed] [Google Scholar]
- 5.Eckenwalder JE (1996) Systematics and evolution of Populus In: Stettler RF, Bradshaw HD, Heilman PE, Hinckler TM editors.Biology of Populus and its implications for management and conservation. Ottawa: Canadian Government Publishing. pp. 7: : 32. [Google Scholar]
- 6.Wu ZY (1999) Flora of China. Volume 20, Fascicule 2. Science Press. pp. 7–78. [Google Scholar]
- 7. Leskinen E, Alström-Rapaport C (1999) Molecular phylogeny of Salicaceae and closely related Flacourtiaceae: Evidence from 5.8 S, ITS 1 and ITS 2 of the rDNA. Plant Systematics and Evolution 215: 209–227. [Google Scholar]
- 8. Smith RL, Sytsma KJ (1990) Evolution of Populus nigra (sect. Aigeiros): introgressive hybridization and the chloroplast contribution of Populus alba (sect. Populus). American journal of botany 1176–1187. [Google Scholar]
- 9. Cervera M, Storme V, Soto A, Ivens B, Van Montagu M, et al. (2005) Intraspecific and interspecific genetic and phylogenetic relationships in the genus Populus based on AFLP markers. Theoretical and Applied Genetics 111: 1440–1456. [DOI] [PubMed] [Google Scholar]
- 10. Hamzeh M, Dayanandan S (2004) Phylogeny of Populus (Salicaceae) based on nucleotide sequences of chloroplast trnT-trnF region and nuclear rDNA. American journal of botany 91: 1398–1408. [DOI] [PubMed] [Google Scholar]
- 11.Slavov GT, Zhelev P (2010) Salient biological features, systematics, and genetic variation of Populus In: Jansson S, Bhalerao R, Groover A, editors. Genetics and Genomics of Populus. Berlin: Springer. pp. 15–38. [Google Scholar]
- 12.Zsuffa L (1975) A summary review of interspecific breeding in the genus Populus L. In: Proceedings of the 14th annual meeting of the Canadian Tree Improvement Association, part 2, 107–123. Canadian Forest Service, Ottawa, Ontario, Canada.Willing R and Pryor L [Google Scholar]
- 13. Willing RR, Pryor LD (1976) Interspecific hybridisation in poplar. Theoretical and Applied Genetics 47: 141–151. [DOI] [PubMed] [Google Scholar]
- 14. Mejnartowicz M (1991) Inheritance of chloroplast DNA in Populus . Theoretical and Applied Genetics 82: 477–480. [DOI] [PubMed] [Google Scholar]
- 15. Page RDM (2000) Extracting species trees from complex gene trees: reconciled trees and vertebrate phylogeny. Molecular Phylogenetics and Evolution 14: 89–106. [DOI] [PubMed] [Google Scholar]
- 16. Duarte JM, Wall PK, Edger PP, Landherr LL, Ma H, et al. (2010) Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels. BMC Evolutionary Biology 10: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bobowski B, Hole D, Wolf P, Bryant L (1999) Identification of roots of woody species using polymerase chain reaction (PCR) and restriction fragment length polymorphism (RFLP) analysis. Molecular Ecology 8: 485–491. [DOI] [PubMed] [Google Scholar]
- 18. Demesure B, Sodzi N, Petit R (1995) A set of universal primers for amplification of polymorphic noncoding regions of mitochondrial and chloroplast DNA in plants. Molecular Ecology 4: 129–134. [DOI] [PubMed] [Google Scholar]
- 19. Huang SS, Hwang SY, Lin TP (2002) Spatial pattern of chloroplast DNA variation of Cyclobalanopsis glauca in Taiwan and East Asia. Molecular Ecology 11: 2349–2358. [DOI] [PubMed] [Google Scholar]
- 20. Shaw J, Lickey EB, Beck JT, Farmer SB, Liu W, et al. (2005) The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. American journal of botany 92: 142–166. [DOI] [PubMed] [Google Scholar]
- 21. Shaw J, Lickey EB, Schilling EE, Small RL (2007) Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. American journal of botany 94: 275–288. [DOI] [PubMed] [Google Scholar]
- 22. Doyle JJ (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19: 11–15. [Google Scholar]
- 23. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic acids research 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic acids symposium series 41: 95–98. [Google Scholar]
- 25. Kelchner SA (2000) The evolution of non-coding chloroplast DNA and its application in plant systematics. Annals of the Missouri Botanical Garden 87: 482–498. [Google Scholar]
- 26. Zhu Q, Ge S (2005) Phylogenetic relationships among A-genome species of the genus Oryza revealed by intron sequences of four nuclear genes. New Phytologist 167: 249–265. [DOI] [PubMed] [Google Scholar]
- 27. Simmons MP, Ochoterena H (2000) Gaps as characters in sequence-based phylogenetic analyses. Systematic Biology 49: 369–381. [PubMed] [Google Scholar]
- 28.Borchsenius F (2009) FastGap 1.2. Department of Biological Sciences, University of Aarhus, Aarhus, Denmark. [Google Scholar]
- 29.Swofford DL (2003) PAUP*: phylogenetic analysis using parsimony, version 4.0 b10. for Macintosh. Sinauer, Sunderland, Massachusetts, USA. [Google Scholar]
- 30. Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 783–791. [DOI] [PubMed] [Google Scholar]
- 31. Posada D (2008) jModelTest: phylogenetic model averaging. Molecular Biology and Evolution 25: 1253–1256. [DOI] [PubMed] [Google Scholar]
- 32. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 33. Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rambaut A (2008) FigTree v1.1.1. Published by the author. [Google Scholar]
- 35. Shimodaira H, Hasegawa M (1999) Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Molecular Biology and Evolution 16: 1114–1116. [Google Scholar]
- 36. Shimodaira H, Hasegawa M (2001) CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17: 1246–1247. [DOI] [PubMed] [Google Scholar]
- 37. Hamzeh M, Périnet P, Dayanandan S (2006) Genetic Relationships among species of Populus (Salicaceae) based on nuclear genomic data. The Journal of the Torrey Botanical Society 133: 519–527. [Google Scholar]
- 38. Chase MW, Zmarzty S, Lledó MD, Wurdack KJ, Swensen SM, et al. (2002) When in doubt, put it in Flacourtiaceae: a molecular phylogenetic analysis based on plastid rbcL DNA sequences. Kew Bulletin 57: 141–181. [Google Scholar]
- 39. Manchester SR, Dilcher DL, Tidwell WD (1986) Interconnected reproductive and vegetative remains of Populus (Salicaceae) from the middle Eocene Green River Formation, northeastern Utah. American journal of botany 156–160. [DOI] [PubMed] [Google Scholar]
- 40. Quanliang S, Qiang Z, Minren H, Mingxiu W (2001) Phologenetic relationship of Populus sections by ITS sequnece analysis. Acta Botanaca Sinica 43: 323–325. [Google Scholar]
- 41. Morin NR (2000) Flora of North America. Flora of North America Association. Volume 7: 5–22. [Google Scholar]
- 42. Levsen ND, Tiffin P, Olson MS (2012) Pleistocene speciation in the genus Populus (Salicaceae). Systematic Biology 61: 401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Manchester SR, Judd WS, Handley B (2006) Foliage and fruits of early poplars (Salicaceae: Populus) from the Eocene of Utah, Colorado, and Wyoming. International Journal of Plant Sciences 167: 897–908. [Google Scholar]
- 44. Barnes BV (1967) Indications of possible mid-Cenozoic hybridization in the aspens of the Columbia Plateau. Rhodora 69. [Google Scholar]
- 45. Zhang D, Zhang Z, Yang K, Li B (2004) Genetic mapping in (Populus tomentosa×Populus bolleana) and P. tomentosa Carr. using AFLP markers. Theoretical and Applied Genetics 108: 657–662. [DOI] [PubMed] [Google Scholar]
- 46. Kuan-yu L, Min-ren H, Ming-xiu W (1997) Study on Origin of Populus tomentosa Carr. Acta Phytotaxonomica Sinica 35: 1. [Google Scholar]
- 47. Rajora O, Dancik B (1995) Chloroplast DNA variation in Populus. II. Interspecific restriction fragment polymorphisms and genetic relationships among Populus deltoides, P. nigra, P. maximowiczii, and P. x canadensis . Theoretical and Applied Genetics 90: 324–330. [DOI] [PubMed] [Google Scholar]
- 48. Eckenwalder JE (1984) Natural intersectional hybridization between North American species of Populus (Salicaceae) in sections Aigeiros and Tacamahaca. II. Taxonomy. Canadian Journal of Botany 62: 325–335. [Google Scholar]
- 49.Little EL Jr, Viereck LA (1971) Conifers and important hardwoods. In: United States. Forest Service, Little EL Jr., Viereck LA, editors. Atlas of United States trees. Washington: Miscellaneous Publications United States Department of Agriculture. Volume 1. [Google Scholar]
- 50. Talbot P, Thompson SL, Schroeder W, Isabel N (2011) An efficient single nucleotide polymorphism assay to diagnose the genomic identity of poplar species and hybrids on the Canadian prairies. Canadian Journal of Forest Research 41: 1102–1111. [Google Scholar]
- 51. Hamzeh M, Sawchyn C, Perinet P, Dayanandan S (2007) Asymmetrical natural hybridization between Populus deltoides and P. balsamifera (Salicaceae) This note is one of a selection of papers published in the Special Issue on Poplar Research in Canada. Botany 85: 1227–1232. [Google Scholar]
- 52. Thompson SL, Lamothe M, Meirmans PG, Perinet P, Isabel N (2010) Repeated unidirectional introgression towards Populus balsamifera in contact zones of exotic and native poplars. Molecular Ecology 19: 132–145. [DOI] [PubMed] [Google Scholar]
- 53.Eckenwalder JE (1977) Systematics of Populus L.(Salicaceae) in southwestern North America with special reference to sect. Aigeiros Duby. Ph.D. dissertation, University of California, Berkeley, California, USA.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Combined supporting information file. File S1. The information of species (names, altitude, longitude, latitude, collector, collection number and place of voucher deposition). File S2. The information of primers. File S3. The information of nuclear and plastid DNA sequences. File S4. Results of the Shimodaira-Hasegawa test that are significant incongruence (P<0.05). File S5. Phylogenetic trees reconstructed based on plastid fragments using parsimony, most likelihood and Bayesian inference methods. Numbers next to nodes indicated bootstrap support value or posterior probabilities. File S6. Phylogenetic trees reconstructed based on nuclear DNA fragments using parsimony, most likelihood and Bayesian inference methods. Numbers next to nodes indicated bootstrap support value or posterior probabilities.
(RAR)