Abstract
AIM: To investigate the early viral kinetics and interleukin-28B (IL28B) polymorphisms of hepatitis C genotype 6 during pegylated interferon and ribavirin therapy.
METHODS: Sixty-five patients with chronic hepatitis C virus (HCV) infection treated with pegylated interferon and ribavirin (PEG-IFN/RBV) were included, of whom 15 (23.1%), 16 (24.6%) and 34 (52.3%) patients were infected with hepatitis C genotype 1 (HCV-1), genotype 3 (HCV-3) and genotype 6 (HCV-6), respectively. Serum HCV-RNA levels were measured frequently during the first 4-wk of therapy. DNA extracted from samples was analyzed for the IL28B single nucleotide polymorphism (SNP) rs12979860 by polymerase chain reaction and direct sequencing.
RESULTS: During the first 4-wk of therapy, the mean viral decline for patients with HCV-6 (5.55 ± 1.82 log10IU/mL) was comparable to that of patients with HCV-3 (5.55 ± 1.82 log10IU/mL vs 5.86 ± 1.02 log10IU/mL, P = 0.44) and was significantly higher than patients with HCV-1 (5.55 ± 1.82 log10IU/mL vs 4.23 ± 1.99 log10IU/mL, P = 0.04). In the HCV-6 group, the first phase (days 0-2) viral decline was significantly higher in patients with the favorable rs12979860 CC than non-CC genotypes (2.46 ± 1.01 log10IU/mL/wk vs 1.70 ± 0.67 log10IU/mL, respectively, P = 0.045). A statistically insignificant decrease in the second-phase (days 7-28) decline was also found in patients with the CC genotype than those with the non-CC genotype, though not significantly different (1.24 ± 0.64 log10IU/mL/wk vs 0.80 ± 0.65 log10IU/mL/wk, respectively, P = 0.172). At baseline, the SNP genotype was an independent predictor of rapid virological response but not of sustained virological response.
CONCLUSION: The IL28B genotype was linked to an impact on early viral kinetics in response to PEG-IFN/RBV therapy in HCV-6 infected patients.
Keywords: Hepatitis C virus, Genotype 6, Interleukin-28B, rs12979860, Early viral kinetics
Core tip: Assessment of the early viral kinetics of hepatitis C virus (HCV) RNA during pegylated interferon/ribavirin therapy correlates with a specific interleukin-28B polymorphism, potentially revealing insights about the virus-host dynamics. This is the first study to report that single nucleotide polymorphism rs12979860 is well correlated with early viral kinetics in the response to antiviral therapy and leads to a higher rate of rapid virological response in HCV-6 infected patients.
INTRODUCTION
Chronic hepatitis C virus (HCV) infection is a major cause of cirrhosis, end-stage liver disease and hepatocellular carcinoma (HCC)[1]. HCV has been classified into seven major genotypes and several subtypes, which exhibit distinctive patterns of geographic distribution[2,3]. In Thailand, approximately 2.2% of the general population has been chronically infected with HCV and the most common genotypes are HCV genotype 3 (HCV-3), genotype 1 (HCV-1) and genotype 6 (HCV-6), respectively[4]. The current standard therapy for all HCV genotypes is a combination of pegylated interferon (PEG-IFN) and ribavirin (RBV). A sustained virological response (SVR) following antiviral treatment can delay disease progression and decrease HCC development. In the Asian population, treatment with the combined therapy results in different SVR depending on the HCV genotype (50%-60% in HCV-1 infected patients, 80%-90% in HCV-3 infected patients, and 70%-80% in HCV-6 infected patients)[5].
Early viral kinetics as measured by HCV RNA levels during the first few weeks of treatment has been shown to be an important predictor of PEG-IFN-based therapy. Typically, HCV kinetics exhibits a biphasic pattern[6-8]. The first phase is characterized by a decline in serum HCV RNA from days 0-2, which represents the antiviral action of interferon (IFN) in blocking HCV production. The second phase, defined by the reduction of HCV RNA levels during days 7-28, is thought to correlate with clearance of HCV. However, different HCV genotypes exhibit different viral kinetics[9,10]. Moreover, it has been shown that polymorphisms near interleukin-28B (IL28B) gene are strongly associated with early viral kinetics during PEG-IFN/RBV therapy in patients with chronic HCV-1 infection[11-14]. However, this correlation has not been investigated in HCV-6 infected individuals. This study aims to determine the early viral kinetics in HCV-6 infected patients undergoing PEG-IFN / RBV treatment and IL28B polymorphisms. Results were compared to those of patients infected with HCV-1 and HCV-3.
MATERIALS AND METHODS
Patients
We analyzed data from a cohort of patients recruited at our center (King Chulalongkorn Memorial Hospital, Bangkok, Thailand) as described previously[15]. Briefly, 66 treatment-naïve adults with chronic HCV infection received PEG-IFN-α2a (Pegasys, Roche) 180 μg/wk plus weight-based RBV (Copegus, Roche) according to the following body weights: ≤ 75 kg, 1000 mg/d; and > 75 kg, 1200 mg/d. Patients infected with HCV-1 and HCV-3 were treated for a fixed duration of 48 and 24 wk, respectively. Patients infected with HCV-6 who achieved RVR were assigned to treatment for 24 wk and the remaining patients were treated for 48 wk. One patient who lost follow up was excluded from the current cohort. The study protocol had been approved by the Institutional Review Board of the Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand (IRB No. 511/51), and all participants had provided written informed consent. The study followed the Helsinki Declaration and Good Clinical Practice guidelines.
HCV RNA kinetics
Serum HCV RNA levels were assessed at days 0, 2, 7, 14, 21 and 28 by real-time quantitative reverse transcription-polymerase chain reaction (RT-PCR)(COBAS TaqMan HCV assay; Roche Diagnostics, Basel, Switzerland), according to the manufacturer’s instructions. The first phase decline was characterized as the difference in HCV RNA levels (log10 IU/mL) from day 0 to 2. The second phase decline was defined as the best fit slope of serum HCV RNA (log10 IU/mL/wk) levels from days 7 to 28[16]. Rapid virological response (RVR) was defined as undetectable HCV RNA at week 4, while SVR was defined as undetectable HCV RNA 24 wk after the end of treatment.
Single nucleotide polymorphism genotyping
Genomic DNA of HCV patients was extracted from 100 μL of peripheral blood mononuclear cells (PBMC) using the QIAamp DNA Mini Kit according to the manufacturer’s instruction (Qiagen, Germany). The single nucleotide polymorphism (SNP); rs12979860 was identified by PCR using specific primers and direct nucleotide sequencing.
The PCR mixture for rs12979860 amplification consisted of 3 μL of extracted DNA, 5 pmole of each primer including rs12979860_R341 (5’-CTCTTCCTCCTGCGGGACAAG-3’) and rs12979860_F706 (5’-TACACCCGTTCCTGTCCCAAG-3’), 12.5 μL of 2 × Perfect Tag Plus MasterMix (5 PRIME, Gaithersburg, MD) and distilled water to a final volume of 25 μL. After an initial denaturation at 94 °C for 3 min, 40 cycles of amplification were performed, each including denaturation at 94 °C for 30 s, annealing at 62 °C for 30 s and extension at 72 °C for 30 s, then followed by a final extension step at 72 °C for 7 min.
In some cases with insufficient amount of PBMC, nested PCR was performed with samples extracted from plasma. The reaction mixture and amplification condition were the same as described above. The outer set of primers used for the first round of PCR included rs12979860_F (5’-GGCGCTGAGGGACCGCTACGTAAGTCACCG-3’) and rs12979860_R (5’-CGCTGCCCCCAGCTCAGCGCCTCTTCCTCC-3’). Then, 1 μL of the first PCR product was combined with primers including rs12979860_R341 and rs12979860_F706 for the subsequent nested PCR. In order to ensure no contamination occurred, negative control was included in both first round and nested PCR amplification in each experiment. The amplification fragment was separated in 2% agarose gel electrophoresis, stained with ethidium bromide and visualized under UV transilluminator. The PCR products were then purified using GelExtract Mini Kits (5 PRIME, Gaithersburg, MD) and subjected to bidirectional sequencing (First BASE Laboratories, Malaysia) using both forward and reverse primers.
The SNP typing of rs12979860 was analyzed based on the chromatograms of nucleotide bases at the SNP position compared with the reference sequence retrieved from Genbank database (http://www.ncbi.nlm.nih.gov/). The chromatograms were visualized and analyzed using Chromas LITE (v.2.01) and SeqMan (DNASTAR, Medison, WI). The superimposed chromatogram signals at the SNP position was interpreted as heterozygous genotype[17]. According to rs12979860 genotypes, CC was defined as major alleles, CT was defined as heterozygous alleles and TT was defined as minor alleles. In samples that the interpretations using sequencing-based assays were not clear, the results were validated by TaqMan SNP Genotyping Assays (Assay ID AH8823E, Applied Biosystems) based on real-time PCR method as described previously[18].
Statistical analysis
The Mann-Whitney U test or Student’s test were used to compare continuous variables, and the χ2 test or Fisher’s exact test were used to compare categorical variables. HCV-RNA kinetic throughout the first 4 wk of therapy was analyzed according to HCV genotypes. The effect of the IL28B genotypes (CC vs non-CC) on HCV viral decline in HCV-1, HCV-3 and HCV-6 patients was also analyzed. Univariate and multivariate logistic regression was used to assess odd ratios associated with RVR and SVR. All statistical analyses were performed using the SPSS software for Windows version 17.0 (SPSS, Chicago, IL, United States). P < 0.05 for a two-tailed test was considered statistically significant.
RESULTS
Baseline characteristics and SNP rs12979860
Among 65 patients naïve to PEG-IFN/RBV included in this study, there were 15 patients with HCV-1, 16 patients with HCV-3 and 34 patients with HCV-6. Table 1 summarizes baseline characteristics and rs12979860 genotypes of these patients based on HCV genotypes. There were no significant differences in the baseline characteristics between each group with regards to gender distribution, mean age, body mass index (BMI), ALT level, HCV RNA level and the degree of liver fibrosis assessed by histology. The distribution of rs12979860 genotypes (CC, CT and TT) in the entire cohort was 50 (76.9%), 10 (15.4%) and 5 (7.7%), respectively. There was no significant difference in the distribution of the IL28B genotypes between HCV genotypes in this study.
Table 1.
Clinical characteristics and single nucleotide polymorphism rs12979860 of the patients according to hepatitis C virus genotypes
| Characteristics | Genotype 1 | Genotype 3 | Genotype 6 | P value |
| (n = 15) | (n = 16) | (n = 34) | ||
| Age (yr) | 46.6 ± 12.9 | 42.8 ± 8.2 | 41.2 ± 8.4 | NS |
| Sex (male) | 53.3% | 81.3% | 67.6% | NS |
| BMI (kg/m2) | 24.2 ± 12.1 | 21.3 ± 5.7 | 23.7 ± 3.7 | NS |
| ALT (U/L) | 86.3 ± 57.8 | 82.6 ± 51.9 | 62.6 ± 54.5 | NS |
| Log10 HCV RNA (IU/mL) | 6.3 ± 0.8 | 6.0 ± 0.8 | 6.5 ± 0.8 | NS |
| Liver fibrosis score | NS | |||
| Score 0-2 | 73.3% | 66.7% | 71.4% | |
| Score 3-4 | 26.7% | 33.3% | 28.6% | |
| SNP rs12979860 | NS | |||
| CC | 66.7% | 75.0% | 82.3% | |
| CT | 20.0% | 18.8% | 11.8% | |
| TT | 13.3% | 6.2% | 5.9% | |
| RVR | 46.7% | 87.5% | 73.5% | 0.039 |
| SVR | 66.7% | 81.3% | 76.5% | NS |
Data described as mean ± SD or proportions (%). ALT: Alanine aminotransferase; RVR: Rapid virological response; SVR: Sustained virological response; BMI: Body mass index; HCV: Hepatitis C virus; NS: Not significant
RVR were achieved in 87.5% of patients with HCV-3 and 73.5% of patients with HCV-6, which was statistically more significant than that of patients with HCV-1 (46.7%) (P = 0.039). The overall rate of SVR in patients with HCV-3 (81.3%) was higher than that of patients with HCV-6 (76.5%) and was higher than patients with HCV-1 (66.7%), although there was no significant difference (P = 0.627). Among patients who attained RVR, SVR was achieved in 90% of patients with HCV-1, 92.9% of patients with HCV-3 and 88% of patients with HCV-6.
Viral decline according to HCV genotypes
The decreases in HCV RNA level during the first 4 wk were significantly correlated to HCV genotypes. We found that the magnitude of viral decline for patients infected with HCV-1 was lower than those infected with HCV-3 or HCV-6 at all time points. After 4 wk of treatment, the HCV RNA levels had declined by a mean of 4.23 ± 1.99 log10IU/mL in patients infected with HCV-1 compared with 5.86 ± 1.02 and 5.55 ± 1.82 log10IU/mL in those infected with HCV-3 and HCV-6 (P = 0.007 and P = 0.040, respectively). There was no significant difference between patients infected with HCV-3 and HCV-6 (P = 0.437). The mean HCV-RNA levels (log10IU/mL) at different time points during the first 4 wk of PEG-IFN/RBV treatment according to HCV genotypes are shown in Figure 1.
Figure 1.
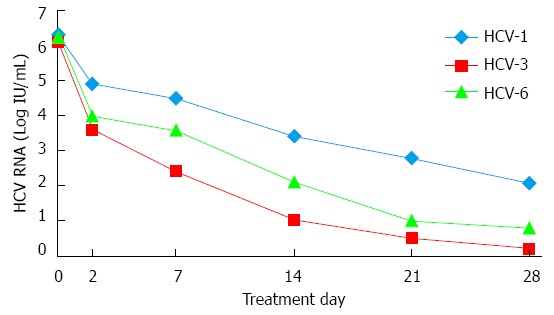
Kinetics of hepatitis C virus based on different viral genotypes. Mean hepatitis C virus (HCV)-RNA levels (log10IU/mL) at different time points (days 0, 2, 7, 14, 21, 28). During the first 4 wk of pegylated interferon and ribavirin therapy according to HCV genotypes. HCV-1: HCV genotype 1; HCV-3: HCV genotype 3; HCV-6: HCV genotype 6.
The HCV-3 and HCV-6 groups had a first phase decline of 2.30 ± 1.25 and 2.32 ± 0.99 log10IU/mL, respectively, which did not achieve statistical difference (P = 0.958). However, both were higher than that of the HCV-1 group (1.38 ± 0.64 log10IU/mL) (P = 0.015 and P = 0.002, respectively). The second phase decline was comparable between the HCV-3 and HCV-6 groups were similar (1.25 ± 0.66 log10IU/mL/wk vs 1.17 ± 0.65 log10IU/mL/wk, respectively, P = 0.675) and higher than that of the HCV-1 group (0.99 ± 0.73 log10IU/mL/wk). The differences between HCV-3 and HCV-1 groups, or between HCV-6 and HCV-1 groups, were not statistically significant (P = 0.310 and P = 0.428, respectively).
Viral decline according to SNP rs12979860
Our data showed that the decreases in HCV RNA level during the first 4 wk of treatment were significantly correlated with the rs12979860 genotypes. Irrespective of viral genotypes, the baseline HCV RNA level in patients with CC genotype was significantly higher than in patients with the non-CC (CT and TT) group (6.42 ± 0.83 and 5.87 ± 0.88 log10IU/mL, respectively, P = 0.040). After 4 wk of treatment, the HCV RNA levels had declined by a mean of 5.77 ± 1.44 log10IU/mL in patients carrying CC compared with 3.84 ± 2.08 log10IU/mL in those carrying non-CC genotype (P < 0.001).
In the HCV-1 group, the HCV RNA levels had declined after 4 wk of treatment by a mean of 5.34 ± 1.43 log10IU/mL in patients carrying CC compared with 2.03 ± 0.50 log10IU/mL in those carrying non-CC genotype (P < 0.001). In the HCV-3 group, the corresponding figures were 5.97 ± 1.01 and 5.23 ± 1.10 log10IU/mL, respectively (P = 0.504). In the HCV-6 group, the corresponding figures were 5.83 ± 1.60 and 4.23 ± 2.34 log10IU/mL, respectively (P = 0.163). The mean HCV-RNA levels (log10IU/mL) at different time points during the first 4 wk of PEG-IFN/RBV treatment according to IL28B genotypes in each HCV genotype are shown in Figure 2.
Figure 2.
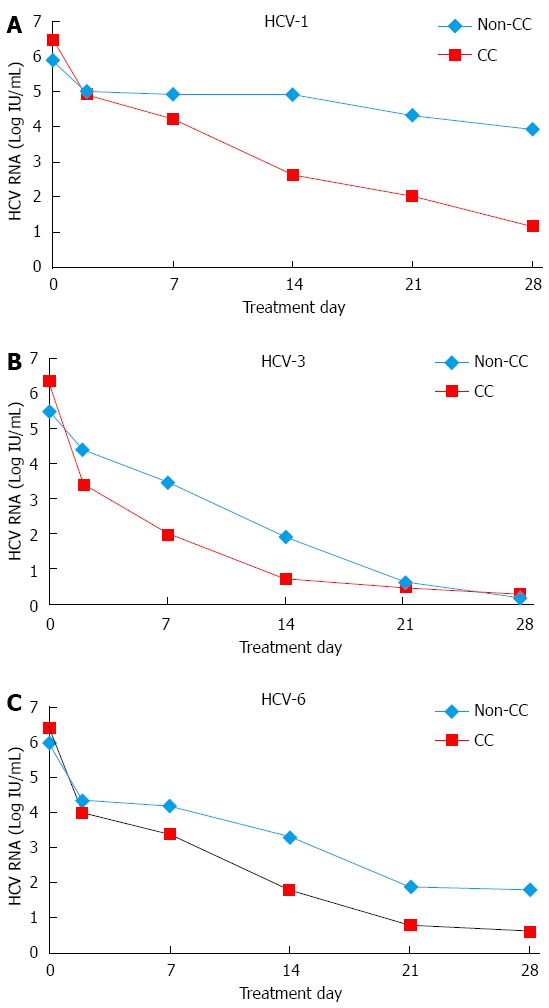
Kinetics of hepatitis C virus based on the single nucleotide polymorphism rs12979860 alleles. Mean hepatitis C virus (HCV)-RNA levels (log10IU/mL) at different time points (days 0, 2, 7, 14, 21, 28). During the first 4 wk of pegylated interferon and ribavirin treatment according to rs12979860 genotypes. A: HCV genotype 1; B: HCV genotype 3; C: HCV genotype 6.
Irrespective of viral genotypes, the first phase decline was significantly higher in patients with the CC than those with non-CC genotype (2.35 ± 1.04 and 1.28 ± 0.63 log10IU/mL, respectively, P < 0.001) (Figure 3A). In addition, the second phase decline was significantly higher for patients with the CC than for those with non-CC genotype (1.27 ± 0.67 and 0.75 ± 0.52 log10IU/mL/wk, respectively, P = 0.004) (Figure 3B). Patients with CC genotype had RVR rates of 78%, whereas RVR in the non-CC group was substantially lower (46.7%, P = 0.027). However, the SVR rates between patients with CC and non-CC genotypes were not significantly different (76.0% and 73.3%, respectively, P = 0.833).
Figure 3.
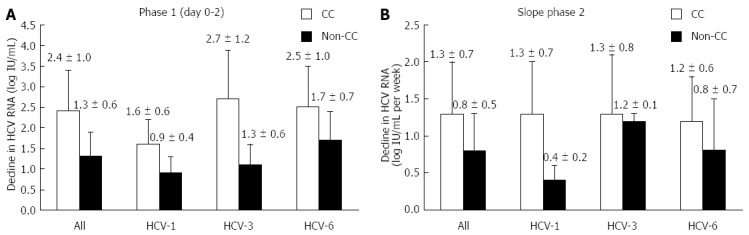
Changes in serum hepatitis C virus RNA concentrations according to rs12979860 genotypes. A: The first phase (days 0-2); B: The second phase (days 7-28) hepatitis C virus (HCV)-1: HCV genotype 1; HCV-3: HCV genotype 3; HCV-6: HCV genotype 6; Data described as mean ± SD.
In the HCV-6 group, the first phase decline was 2.46 ± 1.01 and 1.70 ± 0.67 log10IU/mL in patients with CC and non-CC genotypes, respectively (P = 0.045) (Figure 3A). The second phase decline was 1.24 ± 0.64 and 0.80 ± 0.65 log10IU/mL per week, respectively, in patients with the corresponding IL28B genotypes (P = 0.172) (Figure 3B). Patients with the favorable CC genotype achieved significantly higher RVR rates in comparison to those with the unfavorable genotypes (82.1% and 33.3%, respectively, P = 0.031). However, the SVR rates between patients with CC and non-CC genotypes were not significantly different (78.6% and 66.7%, respectively, P = 0.609).
Factors associated with RVR and SVR in patients with HCV-6
To identify factors associated with RVR and SVR in patients with HCV-6, baseline characteristics of patients and early viral kinetics during therapy were analyzed by univariate and multivariate logistic regression analyses. Potential baseline predictors of RVR and SVR included sex, age, BMI, ALT level, liver fibrosis score, HCV RNA level and rs12979860 genotype.
For RVR, univariate analysis identified significant effects correlated with the presence of the favorable CC genotype (OR = 9.20; 95%CI: 1.30-64.90, P = 0.026), the first phase decline (OR = 3.76; 95%CI: 1.29-10.96, P < 0.015) and second phase kinetics (OR = 9.50; 95%CI: 1.35-66.99, P = 0.024). However, only the first and second phase kinetics remained significant in multivariate logistic regression analysis (the first phase: OR = 6.39; 95%CI: 1.18-34.54, P < 0.031, the second phase; OR = 18.15; 95%CI: 1.13-291.50, P < 0.041). If only baseline pre-treatment parameters were included in the multivariate model, CC genotype (OR = 16.88; 95%CI: 1.45-169.5, P = 0.024) was considered significant.
Variables associated with SVR in univariate analysis were low HCV RNA level (OR = 6.75; 95%CI: 1.11-41.00, P = 0.030) and the second phase kinetics (OR = 12.60; 95%CI: 1.93-82.09, P = 0.008). In multivariate logistic regression analysis, only the second phase kinetics was associated with SVR (OR = 12.79; 95%CI: 1.65-99.28, P = 0.015). If only baseline characteristics were included in the multivariate model, no parameter was significant.
DISCUSSION
In the present study, we demonstrated for the first time the kinetics of HCV-6 correlates with IL28B polymorphisms in patients treated with PEG-IFN/RBV therapy. We found that rs12979860 significantly correlated with the early viral elimination during treatment, with a more rapid reduction of HCV RNA levels in patients carrying the favorable CC allele. The rapid first-phase reduction in patients with the CC allele suggests that the polymorphisms might be linked to the antiviral effect of PEG-IFN/RBV in the blocking of the production or release of HCV. A trend towards a steeper second-phase decline was also found in patients with the CC allele, although this was not statistically significant due to sample size. This finding might indicate that the rs12979860 polymorphism is linked to immune-mediated eradication of infected hepatocytes in addition to the antiviral efficiency.
Our data confirmed previous reports that the decrease in HCV RNA level during the first 4 wk was influenced by HCV genotypes with respect to IL28B polymorphisms. Previous data reported that in patients infected with HCV-1, HCV RNA levels declined by 3.8 log10 IU/mL after 4 wk among patients carrying CC compared with 1.1-1.5 log10IU/mL in patients carrying non-CC allele[19]. In our study, the decrease observed in the HCV-1 group was approximately 5.3 and 2.0 log10IU/mL for those carrying CC and non-CC allele, respectively. Among patients infected with HCV-3, the corresponding declines in this study were 5.97 and 5.23 log10IU/mL, respectively. These observations are in agreement with previous results[20]. In the HCV-6 group, the corresponding figures were 5.83 and 4.23 log10IU/mL, for patients with CC and non-CC allele, respectively. We found that the impact of rs12979860 favorable allele on viral reduction during therapy in patients infected with HCV-6 is at an intermediate level compared to those infected with HCV-3 and HCV-1.
In the HCV-6 group, the collective effects of the SNPs on the first- and second-phase viral decline might explain the differences in RVR rates. For instance, approximately 80% of patients with CC allele and 30% of non-CC allele achieved RVR. However, a high likelihood of RVR in patients carrying the favorable CC allele was not associated with a higher SVR rate. As shown in this study, there was no significant difference in SVR rate between CC and non-CC allele (approximately 78% vs 67%, respectively). In the multivariate model, our data also confirmed that the SNP rs12979860 was associated with RVR but not SVR. Previous studies in HCV-3 cohorts also showed that RVR but not SVR was influenced by the SNP alleles[12,21,22]. In contrast, previous reports in HCV-1 infected patients demonstrated that more robust early declines in serum HCV RNA predicted a higher SVR rate during PEG-IFN/RBV therapy[23]. A possible explanation of the discrepancy among HCV genotypes is that the effect of the SNPs might be attenuated by a better treatment response in HCV-3 or HCV-6 infection. In general, the lowest SVR rate of combined PEG-IFN/RBV therapy is found in HCV-1 infected patients. A much higher SVR rate is observed among HCV-3 infected patients, while the response rate of HCV-6 infection is at an intermediate level[24]. These results suggest that the underlying mechanisms, which link the host genetic status to early HCV suppression, might be different from those required for virus eradication.
In summary, our results demonstrated that SNP rs12979860 was linked to early viral kinetics in response to PEG-IFN/RBV therapy in HCV-6 infected patients, which might lead to a higher rate of RVR. Despite the recent introduction of direct antiviral agents in patients with HCV-1 infection, combined PEG-IFN/RBV therapy is the only currently approved treatment for HCV-6 infection. Thus, we believe that an improved ability to predict treatment response is an important goal and is worthy of further large-scale studies, particularly in south China and many south-east Asian countries in which HCV-6 genotype is prevalent.
ACKNOWLEDGMENTS
We would like to thank the Research Unit of Hepatitis and Liver Cancer and the Center of Excellence in Clinical Virology, Chulalongkorn University, King Chulalongkorn Memorial Hospital and National Research Council of Thailand.
COMMENTS
Background
The current standard therapy for hepatitis C virus (HCV) genotype 6 (HCV-6) is a combination of pegylated interferon (PEG-IFN) and ribavirin (RBV). However, early viral kinetics of HCV RNA levels during the combined therapy in HCV-6 infected patients with regard to interleukin-28B (IL28B) polymorphisms is totally unknown.
Research frontiers
Recently, several genome wide association studies have reported associations of different single nucleotide polymorphisms (SNPs) in close proximity to IL-28B gene with response to antiviral therapy. In addition, several independent studies have confirmed an association between the most studied SNP rs12979860 (located 3 kb upstream of the IL28B gene) and sustained virological response with PEG-IFN/RBV therapy in HCV genotype 1 (HCV-1) infection. In contrast, the importance of the SNP in HCV-6 infection is less defined.
Innovations and breakthroughs
This is the first study to report the early viral kinetics of HCV-6 during treatment in correlation with IL28B polymorphisms. The data demonstrated that rs12979860 CC genotype correlated with the rapid early viral elimination during treatment, particularly during the first phase. In addition, the impact of the SNP genotypes on viral reduction during therapy in patients infected with HCV-6 is at an intermediate level between those infected with HCV genotype 3 (HCV-3) and HCV-1.
Applications
SNP rs12979860 was linked to an impact on early viral kinetics in response to PEG-IFN/RBV therapy in HCV-6 infected patients, which leads to a higher rate of rapid virological response. This information may be useful for clinicians in counseling individual HCV-6 infected patients who are receiving antiviral therapy, particularly in south China and many south-east Asian countries in which this HCV genotype is highly prevalent.
Terminology
Early viral kinetics of HCV is demonstrated by measurement of HCV RNA levels frequently during the first few weeks of PEG-IFN and RBV treatment. Typically, the early kinetics has a biphasic pattern. The first phase is characterized by a decline in serum HCV RNA from days 0-2, which represents the antiviral action of IFN in blocking HCV production. The second phase, defined by the reduction of HCV RNA levels during days 7-28, indicates the clearance of infected cells by immune response.
Peer review
The paper is well written and gives important information of the viral kinetics of HCV-6 infection, which is considered to be a lesser known HCV genotype. Thus, these data may provide a better understanding of HCV-host dynamics in response to antiviral treatment.
Footnotes
Supported by Chulalongkorn University, No. RES560530155; Thailand Research Fund, No. BRG5580005, and No. DPG5480002; Joint Research Program between National Research Council of Thailand and Japan Society for the Promotion of Science, Centenary Academic Development Project, No. CU56-HR01; Ratchadapiseksompotch Fund (Faculty of Medicine), Integrated Innovation Academic Center and Postdoctoral of Ratchadaphiseksomphot Endowment Fund, Chulalongkorn University; Higher Education Research Promotion and National Research University Project of Thailand, Office of the Higher Education Commission, No. HR1155A
P- Reviewer: Al-Shamma S, Rodriguez-Frias F S- Editor: Gou SX L- Editor: A E- Editor: Ma S
References
- 1.Lavanchy D. The global burden of hepatitis C. Liver Int. 2009;29 Suppl 1:74–81. doi: 10.1111/j.1478-3231.2008.01934.x. [DOI] [PubMed] [Google Scholar]
- 2.Argentini C, Genovese D, Dettori S, Rapicetta M. HCV genetic variability: from quasispecies evolution to genotype classification. Future Microbiol. 2009;4:359–373. doi: 10.2217/fmb.09.8. [DOI] [PubMed] [Google Scholar]
- 3.Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology. 2014;59:318–327. doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akkarathamrongsin S, Praianantathavorn K, Hacharoen N, Theamboonlers A, Tangkijvanich P, Tanaka Y, Mizokami M, Poovorawan Y. Geographic distribution of hepatitis C virus genotype 6 subtypes in Thailand. J Med Virol. 2010;82:257–262. doi: 10.1002/jmv.21680. [DOI] [PubMed] [Google Scholar]
- 5.Nguyen LH, Nguyen MH. Systematic review: Asian patients with chronic hepatitis C infection. Aliment Pharmacol Ther. 2013;37:921–936. doi: 10.1111/apt.12300. [DOI] [PubMed] [Google Scholar]
- 6.Neumann AU, Lam NP, Dahari H, Gretch DR, Wiley TE, Layden TJ, Perelson AS. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998;282:103–107. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 7.Colombatto P, Civitano L, Oliveri F, Coco B, Ciccorossi P, Flichman D, Campa M, Bonino F, Brunetto MR. Sustained response to interferon-ribavirin combination therapy predicted by a model of hepatitis C virus dynamics using both HCV RNA and alanine aminotransferase. Antivir Ther. 2003;8:519–530. [PubMed] [Google Scholar]
- 8.Herrmann E, Lee JH, Marinos G, Modi M, Zeuzem S. Effect of ribavirin on hepatitis C viral kinetics in patients treated with pegylated interferon. Hepatology. 2003;37:1351–1358. doi: 10.1053/jhep.2003.50218. [DOI] [PubMed] [Google Scholar]
- 9.Kohara M, Tanaka T, Tsukiyama-Kohara K, Tanaka S, Mizokami M, Lau JY, Hattori N. Hepatitis C virus genotypes 1 and 2 respond to interferon-alpha with different virologic kinetics. J Infect Dis. 1995;172:934–938. doi: 10.1093/infdis/172.4.934. [DOI] [PubMed] [Google Scholar]
- 10.Lindh M, Arnholm B, Eilard A, Färkkilä M, Hellstrand K, Lagging M, Langeland N, Mørch K, Nilsson S, Pedersen C, et al. Hepatitis C treatment response kinetics and impact of baseline predictors. J Viral Hepat. 2011;18:400–407. doi: 10.1111/j.1365-2893.2010.01323.x. [DOI] [PubMed] [Google Scholar]
- 11.Bochud PY, Bibert S, Negro F, Haagmans B, Soulier A, Ferrari C, Missale G, Zeuzem S, Pawlotsky JM, Schalm S, et al. IL28B polymorphisms predict reduction of HCV RNA from the first day of therapy in chronic hepatitis C. J Hepatol. 2011;55:980–988. doi: 10.1016/j.jhep.2011.01.050. [DOI] [PubMed] [Google Scholar]
- 12.Scherzer TM, Hofer H, Staettermayer AF, Rutter K, Beinhardt S, Steindl-Munda P, Kerschner H, Kessler HH, Ferenci P. Early virologic response and IL28B polymorphisms in patients with chronic hepatitis C genotype 3 treated with peginterferon alfa-2a and ribavirin. J Hepatol. 2011;54:866–871. doi: 10.1016/j.jhep.2010.08.024. [DOI] [PubMed] [Google Scholar]
- 13.Lindh M, Lagging M, Arnholm B, Eilard A, Nilsson S, Norkrans G, Söderholm J, Wahlberg T, Wejstål R, Westin J, et al. IL28B polymorphisms determine early viral kinetics and treatment outcome in patients receiving peginterferon/ribavirin for chronic hepatitis C genotype 1. J Viral Hepat. 2011;18:e325–e331. doi: 10.1111/j.1365-2893.2010.01425.x. [DOI] [PubMed] [Google Scholar]
- 14.Stenkvist J, Sönnerborg A, Weiland O. HCV RNA decline in chronic HCV genotype 2 and 3 during standard of care treatment according to IL28B polymorphism. J Viral Hepat. 2013;20:193–199. doi: 10.1111/j.1365-2893.2012.01645.x. [DOI] [PubMed] [Google Scholar]
- 15.Tangkijvanich P, Komolmit P, Mahachai V, Poovorawan K, Akkarathamrongsin S, Poovorawan Y. Response-guided therapy for patients with hepatitis C virus genotype 6 infection: a pilot study. J Viral Hepat. 2012;19:423–430. doi: 10.1111/j.1365-2893.2011.01566.x. [DOI] [PubMed] [Google Scholar]
- 16.Howell CD, Gorden A, Ryan KA, Thompson AJ, Ibrahim C, Fried M, Afdhal NH, McHutchison JG, Shianna KV, Goldstein DB, et al. Single nucleotide polymorphism upstream of interleukin 28B associated with phase 1 and phase 2 of early viral kinetics in patients infected with HCV genotype 1. J Hepatol. 2012;56:557–563. doi: 10.1016/j.jhep.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akkarathamrongsin S, Sugiyama M, Matsuura K, Kurbanov F, Poovorawan Y, Tanaka Y, Mizokami M. High sensitivity assay using serum sample for IL28B genotyping to predict treatment response in chronic hepatitis C patients. Hepatol Res. 2010;40:956–962. doi: 10.1111/j.1872-034X.2010.00702.x. [DOI] [PubMed] [Google Scholar]
- 18.Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson AJ, Muir AJ, Sulkowski MS, Ge D, Fellay J, Shianna KV, Urban T, Afdhal NH, Jacobson IM, Esteban R, et al. Interleukin-28B polymorphism improves viral kinetics and is the strongest pretreatment predictor of sustained virologic response in genotype 1 hepatitis C virus. Gastroenterology. 2010;139:120–9.e18. doi: 10.1053/j.gastro.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 20.Lindh M, Lagging M, Färkkilä M, Langeland N, Mørch K, Nilsson S, Norkrans G, Pedersen C, Buhl MR, Westin J, et al. Interleukin 28B gene variation at rs12979860 determines early viral kinetics during treatment in patients carrying genotypes 2 or 3 of hepatitis C virus. J Infect Dis. 2011;203:1748–1752. doi: 10.1093/infdis/jir193. [DOI] [PubMed] [Google Scholar]
- 21.Stättermayer AF, Stauber R, Hofer H, Rutter K, Beinhardt S, Scherzer TM, Zinober K, Datz C, Maieron A, Dulic-Lakovic E, et al. Impact of IL28B genotype on the early and sustained virologic response in treatment-naïve patients with chronic hepatitis C. Clin Gastroenterol Hepatol. 2011;9:344–350.e2. doi: 10.1016/j.cgh.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 22.Bucci C, von Delft A, Christian A, Flemming VM, Harrison A, Halliday J, Collier J, Manganis C, Klenerman P, Irving W, et al. ‘Favourable’ IL28B polymorphisms are associated with a marked increase in baseline viral load in hepatitis C virus subtype 3a infection and do not predict a sustained virological response after 24 weeks of therapy. J Gen Virol. 2013;94:1259–1265. doi: 10.1099/vir.0.051052-0. [DOI] [PubMed] [Google Scholar]
- 23.Jia Z, Ding Y, Tian S, Niu J, Jiang J. Test of IL28B polymorphisms in chronic hepatitis C patients treated with PegIFN and ribavirin depends on HCV genotypes: results from a meta-analysis. PLoS One. 2012;7:e45698. doi: 10.1371/journal.pone.0045698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang TJ, Ghany MG. Current and future therapies for hepatitis C virus infection. N Engl J Med. 2013;368:1907–1917. doi: 10.1056/NEJMra1213651. [DOI] [PMC free article] [PubMed] [Google Scholar]


