Figure 1.
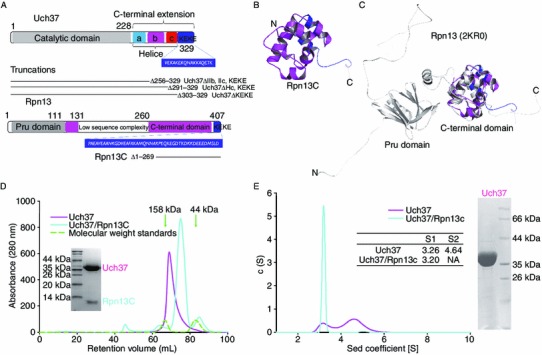
Oligomerization of Uch37 in solution. (A) Diagrammatic representation of the constructs of Uch37 and Rpn13 used in this study. (B) Cartoon representation of the structure of Rpn13C solved by NMR. The color representation of Rpn13 is shown as the same as Fig. 1A. (C) Overlay of the cartoon of the NMR structure of Rpn13C (AA 270–407) over the NMR structure of the full-length Rpn13 (grey color; PDB ID 2KR0). The truncation had no effect on the folding of the protein. (D) Size exclusion profiles revealed that Uch37 has a shorter retention time than the Uch37-Rpn13C complex. Proteins from the peaks were analyzed by SDS-PAGE (inset). Ovalbumin (44 kDa) and γ-globulin (158 kDa) were run as standards under identical conditions. The positions of the standards are marked with green arrows. (E) Analytical ultracentrifugation (AUC) analysis of Uch37 and the Uch37-Rpn13C complex. Uch37 was heterogeneous and exhibited two distinct peaks, whereas the Uch37-Rpn13C complex was homogenous. The sedimentation co-efficient (S value) calculated for Uch37 and the Uch37-Rpn13C complex are shown in the inset
