Abstract
Quantitative real-time PCR (qPCR) is becoming a popular tool for the quantification of gene expression in the brain and endocrine tissues of songbirds. Accurate analysis of qPCR data relies on the selection of appropriate reference genes for normalization, yet few papers on songbirds contain evidence of reference gene validation. Here, we evaluated the expression of ten potential reference genes (18S, ACTB, GAPDH, HMBS, HPRT, PPIA, RPL4, RPL32, TFRC, and UBC) in brain, pituitary, ovary, and testis in two species of songbird: zebra finch and white-throated sparrow. We used two algorithms, geNorm and NormFinder, to assess the stability of these reference genes in our samples. We found that the suitability of some of the most popular reference genes for target gene normalization in mammals, such as 18S, depended highly on tissue type. Thus, they are not the best choices for brain and gonad in these songbirds. In contrast, we identified alternative genes, such as HPRT, RPL4 and PPIA, that were highly stable in brain, pituitary, and gonad in these species. Our results suggest that the validation of reference genes in mammals does not necessarily extrapolate to other taxonomic groups. For researchers wishing to identify and evaluate suitable reference genes for qPCR songbirds, our results should serve as a starting point and should help increase the power and utility of songbird models in behavioral neuroendocrinology.
Keywords: housekeeping gene, quantitative real-time PCR (qPCR), reference gene, songbird, white-throated sparrow, zebra finch
Introduction
Quantitative real-time PCR (qPCR) as a method to measure RNA expression was developed over 15 years ago (VanGuilder et al., 2008). As qPCR technologies and techniques have been refined, the method has increasingly been chosen over alternatives, such as Northern blotting, in situ hybridization, and RNase protection assays, for work in species for which genetic sequence is readily available (reviewed in VanGuilder et al., 2008). qPCR offers a rapid and sensitive way to quantify gene expression when knowing the precise location of that expression within the tissue of interest is not important. Even when the location of the expression is important, for example in specific brain regions, microdissection techniques can be used to prepare samples for qPCR. The technique has been used to link gene expression, hormones, and behavior for almost a decade in rodents (e.g., Levin et al., 2004; Jasnow et al., 2006).
To truly understand the neuroendocrine basis of highly derived social behaviors, we need to choose animal models with rich social repertoires – in other words, the species that most closely model the behaviors we want to study. Advances in genomic technology are making it more and more feasible to bridge from well-characterized data-rich lab animals, such as rats and mice, to phenomena-rich species such as fish, lizards, and songbirds (Clayton & London, 2014; Insel & Fernald, 2004; Robinson et al., 2005; 2008). Songbirds in particular provide valuable model systems in which to study the dynamic relationship between genes, hormones, and behavior because the existing database on avian social behavior is unparalleled. Although songbirds could provide profound insight into the neuroendocrine basis of diverse social behaviors, they have been underutilized by neuroendocrinologists.
Recently, with the advent of highly accessible genomic resources for songbirds (e.g., Replogle et al., 2008; Warren et al., 2010), there has been a dramatic increase in the number of studies designed to elucidate the relationships between gene expression, hormones, and behavior. This increase is partly attributable to the development of a microarray based on zebra finch cDNA as part of the Songbird Neurogenomics (SoNG) initiative (Replogle et al., 2008). In many studies published between 2005 and 2010, qPCR was used to validate microarray results (e.g., Jones et al., 2008a, 2008b; Mukai et al., 2009). After 2010, with the increased availability of genomic sequence from a variety of songbirds, the number of species represented in qPCR studies dramatically increased (Table 1). Overall, qPCR has been used in songbirds to quantify expression of mRNA in relation to stress responses, maternal care, photoperiod, circadian rhythm, migration, aggression, sexual differentiation, and singing behavior. Thus, this technique is already advancing the study of gene expression in songbirds as it has in rodents. As application of the technique expands, it is important that it be appropriately employed for the species or tissue under investigation.
Table 1.
Studies in which qPCR was used to measure gene expression in songbird brain, pituitary, or gonad.
| Reference | Species | Tissuea | Reference gene |
|---|---|---|---|
| Wade et al. (2005) | Taeniopygia guttata | telencephalon | GAPDHb,c |
| Lombardino et al. (2006) | Taeniopygia guttata | HVC projection neurons | normalized to total RNAc |
| Jones et al. (2008a) | Zonotrichia leucophrys gambelii | telencephalon | GAPDHc,d |
| Jones et al. (2008b) | Zonotrichia leucophrys gambelii | telencephalon | GAPDHb,c,d |
| Kim et al. (2008) | Serinus canaria | HVC | GAPDH |
| Wynne et al. (2008) | Taeniopygia guttata | telencephalon | GAPDHb |
| Mukai et al. (2009) | Melospiza melodia | hypothalamus | ACTBc,d |
| Tomaszycki et al. (2009) | Taeniopygia guttata | telencephalon | GAPDHb,c,d |
| Mirzatoni et al. (2010) | Taeniopygia guttata | cerebellum | GAPDH |
| Cerasale et al. (2011) | Zonotrichia albicollis | hypothalmus | ACTB |
| Duncan and Saldanha (2011) | Taeniopygia guttata | telencephalon | GAPDH |
| Banerjee et al. (2012) | Taeniopygia guttata | cerebellum, Hp, hypothalamus | ACTB |
| Perfito et al. (2012) | Parus major | MBH, pituitary | HPRT, GAPDH, PK-a, PK-t |
| Qi et al. (2012) | Taeniopygia guttata | telencephalon | GAPDHb |
| Rosvall et al. (2012) | Junco hyemalis | hypothalamus, PTR, and VmT | GAPDH |
| Stevenson and Ball (2012) | Serinus canaria | MBH | GAPDH |
| Thompson et al. (2012) | Zonotrichia leucophrys gambelii | HVC and RA | 18Sc,d |
| Bentley et al. (2013a) | Sturnus vulgaris | Area X | HPRT, GAPDH |
| Bentley et al. (2013b) | Sturnus vulgaris | MBH | HPRT, GAPDH, PK-a, PK-t, |
| Bergeon Burns et al. (2013) | Junco hyemalis | hypothalamus, PTR, and VmT | GAPDHb |
| Duncan et al. (2013) | Taeniopygia guttata | telencephalon | GAPDH |
| Egbert et al. (2013) | Passer domesticus | ovarian follicles | 18S |
| Liebl and Martin (2013) | Passer domesticus | Hp | 18S |
| Liebl et al. (2013) | Passer domesticus | Hp | 18S |
| Medina et al. (2013) | Passer domesticus | whole brain | GAPDH, TBP |
| Rosvall et al. (2013) | Junco hyemalis | ovary, rostral hypothalamus | GAPDH |
| Shi et al. (2013) | Taeniopygia guttata | Area X | GAPDH, U6b,e |
| Singh et al. (2013) | Emberiza bruniceps | hypothalamus, testes | ACTB |
| Bergeon Burns et al. (2014) | Junco hyemalis | rostral hypothalamus, testes | GAPDH |
Abbreviations: Hp: hippocampus, HVC: used as a proper name, MBH: medial basal hypothalamus, PK: protein kinase, PTR: right posterior telencephalon, RA: robust nucleus of the arcopallium, VmT: ventromedial telencephalon.
Authors verified that GAPDH expression did not vary between experimental groups.
qPCR used to validate microarray findings,
SoNG microarray characterized in Replogle et al. 2008,
U6 used as reference gene for micro-RNAs.
Because small variations due to technical factors can have large effects on experimental outcomes, it is critical that qPCR data be normalized to reduce this variability. The most commonly utilized method, in studies of mammals and songbirds alike, is to normalize gene expression to an internal control, or reference gene (often referred to as a housekeeping gene). Appropriate reference genes should be constitutively and equally expressed in the tissues or cells under investigation and should not change across experimental groups or conditions (e.g., age, sex, hormonal states, photoperiod, treatments) (Andersen et al., 2004, de Jonge et al., 2007, Vandesompele et al., 2002). Because all genes are regulated to some extent, and expression varies by tissue or cell type and experimental conditions, reference genes must be carefully selected and validated based on the experimental model of interest (Bustin et al., 2009). Although a number of reference genes have been validated for normalization in humans and mice, one cannot a priori assume that the same genes will serve as appropriate reference genes in an avian model. For example, expression of the commonly used housekeeping genes 18S ribosomal RNA (18S), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and β-actin (ACTB), has been observed to vary with photoperiod or singing in songbird neural tissue (Bentley et al., 2013b; Lombardino et al., 2006; Perfito et al., 2012; Wada et al., 2006). In a recent study, Medina et al. (2013) quantified mRNA expression of target genes in brain tissue of house sparrows (Passer domesticus) and normalized expression to two common reference genes: TATA-box binding protein (TBP) and GAPDH. They reported that when target gene expression was normalized to TBP, the resulting values were not correlated with values obtained when normalizing to GAPDH. In other words, one or both of the reference genes varied across samples and were “not telling the same story” (p.33). These results highlight the importance of identifying good reference genes and performing proper validation.
In 24 out of 29 papers published between 2005 and 2013 in which qPCR was used to measure gene expression in songbirds, normalization was performed relative to a single reference gene: 18S, GAPDH, or ACTB (Table 1). In only seven of these studies did the authors state that reference gene expression was stable across experimental groups. In three studies (Bentley et al., 2013a, 2013b; Perfito et al., 2012), the authors used geNorm to calculate a normalization factor based on the expression of multiple reference genes, which was used to normalize target gene expression. With an increasing number of researchers wishing to perform gene expression analysis in songbirds, there is a growing need to evaluate the quality of reference genes in these species, and to identify suitable reference genes. Because best practices dictate the use of more than one reference gene (Bustin et al., 2009, Vandesompele et al., 2002), validation of multiple genes in songbirds would be useful.
In this study, we assessed the stability of multiple commonly used reference genes in four tissues: brain, pituitary, ovary, and testis. We chose zebra finch (Taeniopygia guttata) and white-throated sparrow (Zonotrichia albicollis) as two examples of songbird models used in endocrine studies. The zebra finch is a popular model for studying the biological basis of social behavior and sex differences, and was the first songbird for which whole genome sequence became available (Warren et al., 2010). The white-throated sparrow and its congener, the white-crowned sparrow (Z. leucophrys), are popular field endocrine models studied in their natural habitats (Maney, 2008; Wingfield & Farner, 1993). In addition, white-throated sparrows exhibit a plumage and behavioral polymorphism linked to a chromosomal rearrangement (Thorneycroft, 1966; Thorneycroft, 1975), making them ideal for sociogenomic studies (Horton et al., 2014; Maney, 2008).
Our selection of candidate reference genes (Table 2) was based on reports that they are stably expressed in other organisms. These genes (18S, ACTB, GAPDH, HMBS, HPRT, PPIA, RPL4, RPL32, TFRC and UBC) have been validated as suitable reference genes in a variety of organisms and tissues, including human and mouse brain and testis (Boda et al., 2009; Cheng et al., 2011) and chicken blood and fibroblasts (De Boever et al., 2008; Yin et al., 2010; Yue et al.). We also selected reference genes to represent a variety of functional classes to minimize the effects of co-regulation. We measured reference gene stability using two software programs: geNorm and NormFinder (Andersen et al., 2004; Vandesompele et al., 2002).
Table 2.
Reference and target genes evaluated in this study and primer sequences.
| Symbol | Gene Name | Primer Sequencea | Probe | Accession No. or Gene ID |
Function |
|---|---|---|---|---|---|
| 18S | 18S ribosomal RNA | F: gggtgagttttcccgtgtt R: aacttaaaggaattgacggaagg |
131 | HQ873432.1b | cytosolic small ribosome subunit, translation |
| ACTB | Actin, beta | F (ZAL): ccaaccgcgagaagatga F (TGU): ccaaagccaacagagagaaga R: gtggtacgaccagaggcatac |
56 |
XM_002192780.1 NM_205518b |
cytoskeletal structural protein |
| APOD | Apolipoprotein D | F: ctgggggtccgctttaac R: gtcagtggagatgacccagtaa |
40 | XM_002188195.2 | Lipid transporter activity/cholesterol binding |
| CGA | Chorionic gonadotropin alpha polypeptide | F: ccatcctgcagtttctctcatt R: agctgcatacttcccatagca |
94 | XM_005485963.1 | Subunit of glycoprotein hormones |
| FSHR | Follicle stimulating hormone receptor | F: gagaaagccaacaacctcgt R: ccgaaggccagtgtttga |
97 | XM_005491476.1 | Gonadal growth and spermatogenesis |
| GAPD H | Glyceraldehyde-3-phosphate dehydrogenase | F: caacttcggcattgtggag R: ggcccatccactgtcttct |
76 | NM_001198610.1 | glycolysis |
| HMBS | Hydroxymethylbilane synthase | F: ccggtgttagttatccccatc R: gggcctgaccaggaacat |
76 | XM_002187761.1 | heme biosynthesis |
| HPRT | Hypoxanthine guanine phosphoribosyl transferase | F: caatgaatacttcagagatttgaacc R: tgaactctgctttcatgctttg |
38 | XM_002190239.2 | purine synthesis in salvage pathway |
| PPIA | Peptidylprolyl isomerase A (cyclophilin A) | F (ZAL): agaagggatttggctacaagg F (TGU): gaagggcttcggctacaag R: ccattgtggcgtgtgaagt |
38 | NM_001245462 | peptidyl-prolyl cis-trans isomerase activity |
| RPL4 | Ribosomal protein L4 | F: atgagaagccaggaaatcca R (ZAL): cagtgggttcttcttcaggacc R (TGU): cagtgggttcttcttcaggact |
143 | ENSTGUT00000009825 | 60S ribosomal protein subunit, translation |
| RPL32 | Ribosomal protein L32 | F: tgtaaaagagctggaagtgc R: ttcttcgaggacacgttgtg |
136 | NM_001252255.1b | 60S ribosomal protein subunit, translation |
| TFRC | Transferrin receptor (p90, CD71) | F (ZAL): ggcacttcaaagacgatgtg F (TGU): ggcacttcaatgccaatgtg R: tcgatgtctcttctcagtgcat |
11 | ENSTGUT00000009948 | endocytosis, proteolysis |
| UBC | Ubiquitin C | F: gcaccgctggatacattttac R: tcacattttcaatggtgtcactg |
39 | NM_001245834.1 | signal transduction |
F: forward primer, R: reverse primer, ZAL: primer used in white-throated sparrow, TGU: primer used in zebra finch.
Gallus gallus sequences were used to search the zebra finch genome to identify gene sequences not annotated in the zebra finch genome.
Methods and Materials
Tissue collection
All animal procedures were approved by Emory University’s Animal Care and Use Committee. Zebra finches were obtained from a local vender (5 females, 7 males) and from a breeding colony at Georgia State University (6 females, 4 males). All were maintained on a light regime of 12L:12D. Animals were sacrificed by isoflurane overdose. Their brains were rapidly removed, bisected along the midline, frozen on dry ice, and stored at −80°C. Pituitaries and gonads were placed in RNAlater (Life Technologies, Grand Island, NY). One of the pituitaries broke apart during collection and was not available for analysis. One of the females had a large yolky follicle that was removed from the ovary before collection. The ovarian follicles of the other females were no larger than 5mm in diameter. Tissues were incubated in RNAlater at 4°C for 48 hrs, then the RNAlater was removed and samples were stored at −80°C.
White-throated sparrows were collected in mist nets during May and June 2010 and 2011 in the Hemlock Stream Forest in Argyle, Maine, USA (46 males, 33 females). The sparrows were determined to be in one of two breeding stages at the time of collection: pre-parental (collected during the pre-nesting, nest building, and laying phases; 25 males, 13 females) or parental (collected during the nestling phase; 21 males, 20 females). This species exhibits two plumage morphs, white-striped and tan-striped, determined by the presence or absence of a chromosomal rearrangement (Thomas et al., 2008; Throneycroft, 1975). In our sample, morph was confirmed by PCR genotyping (Horton et al., 2013; Michopoulos et al., 2007). Immediately after capture, birds were rapidly sacrificed by isoflurane overdose. Brains were rapidly removed, immediately frozen on dry ice and stored at −80. Pituitaries and gonads were collected as described above.
Total RNA isolation and cDNA synthesis
For the zebra finches, RNA was extracted from one hemisphere of each brain: the left hemisphere for 6 males and 4 females, and the right for 5 males and 7 females. Qiazol (Qiagen, Hilden, Germany) was added directly to the frozen hemispheres and the samples were allowed to thaw for 30 sec. before homogenization. The sparrow brains were cryosectioned at 20µm thickness, and every eighth section was placed into a tube of RNAlater and stored at −80°C. The RNAlater was removed prior to addition of Qiazol solution. All tissue samples were homogenized with a rotor stator homogenizer or pellet mixer. Additional homogenization of brain samples was done using Qiashredders according to manufacturer’s protocol (Qiagen). Total RNA was extracted from brain samples using a miRNeasy kit (Qiagen). Pituitary and gonad RNA was extracted using the High Pure RNA isolation kit (Roche Diagnostics, Indianapolis, IN). On-column DNAse digestion was performed on all samples according to manufacturer’s protocol. Three finch pituitary samples failed to yield sufficient RNA for analysis (2 male, 1 female). The quality of sparrow RNA was assessed on a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) using RNA Pico assays; the quality of RNA samples collected in 2010 was similar to those collected in 2011. Random hexamer primed reverse transcription was performed using a Transcriptor first strand cDNA synthesis kit (Roche). The amount of RNA used per 20μl cDNA reaction was as follows: gonad 200ng, brain 400ng, sparrow pituitary 200ng, and finch pituitary 100ng.
Primer Design
Primers and probe sets were designed using Roche universal primer design software and GenBank or Ensemble sequences (Table 2). For genes not annotated in the zebra finch genome (18S and RPL32), we used chicken sequences to perform a BLAT search of the zebra finch genome (build WUGSC 3.2.4/taeGut1, UCSC genome browser, http://genome.ucsc.edu/). Primers were then designed against zebra finch sequence. The amplicons were then used to search a white-throated sparrow transcriptome (J. W. Thomas, unpublished) to obtain sparrow sequence. Sparrow-specific primers were designed if there was more than one mismatch between sparrow and finch (PPIA and TFRC) or when a mismatch was located at the last base of the primer (RPL4). When the sequences contained no mismatches or a single mismatch at any other location, the same primers were used for both species (Table 2). The primer set designed from the zebra finch ACTB sequence produced an amplicon in sparrow pituitary and gonad, but not sparrow brain or any of the finch tissues. Therefore, an alternate forward ACTB primer for finch samples was designed based on chicken ACTB sequence. Amplification of a single band was verified by running the PCR product on an agarose gel. Amplification efficiencies, calculated using standard curves, were 90–110%. We did not test HMBS in any of the finch tissues or sparrow brain, or UBC in any of the sparrow tissues, because multiple amplicons were observed. Due to limited sample availability, only five genes were run in the sparrow pituitary samples.
Quantitative real-time polymerase chain reaction
(qPCR) qPCR was performed using the Roche LightCycler 480 Real-Time PCR System in combination with Roche Universal ProbeLibrary (UPL) hydrolysis probes. Reactions were run in triplicate in 384 well plates containing 2.5 µl of a 1:20 dilution of cDNA, 2x Probes Master (5 µl, Roche), 0.2 µl UPL probe, and 0.5 µM each forward and reverse primers, in a total reaction volume of 10 µl. Every plate included negative controls consisting of no reverse transcriptase reactions (no RT in the cDNA reaction) and a no template control (water in place of cDNA). Cycling conditions were 95°C for 10 min and 45 cycles of 95°C for 10 sec, 60°C for 30 sec, and 72°C for 1 sec. Crossing point (Cp) values were calculated using the Abs Quant/ 2nd Derivative Max method using LightCycler 480 software. For four of the zebra finch samples (two male brain, one female pituitary, and one ovary), all of the reactions produced unusually low yield; those samples were excluded from subsequent analysis.
Data Analysis
Samples were divided into eight sets based on tissue type (brain, pituitary, ovary, or testis) and species (finch or sparrow). Data from each set were entered into separate geNorm analyses. The geNorm program calculates an expression stability value (M) for each gene based on the average pair-wise variation with all other genes in the analysis. Genes are then ranked from least to most stable (highest to lowest M value). Average expression stability values are calculated from M values of remaining genes during stepwise exclusion of the least stable gene (highest M value). Stepwise exclusion produces a pair of genes with the lowest average M value, which cannot be ranked in order (Vandesompele et al., 2002). For highest accuracy, normalization using multiple reference genes, rather than a single gene, is recommended. To normalize target gene expression to the expression of multiple reference genes, a normalization factor is calculated based on the expression of the most stable genes. The optimal number of reference genes is determined by calculation of the pairwise variation value (Vn/n+1), which reflects the effect of stepwise inclusion of the next most stable gene on the normalization factor. A large pairwise variation means the additional gene significantly impacts the normalization factor and should be included as a reference gene when normalizing target gene data. We used a cutoff value of 0.15 to determine the optimal number of reference genes (Vandesompele et al., 2002).
We also analyzed our data using the NormFinder algorithm, which tests whether reference gene expression is stable across groups (such as sex) or experimental conditions. NormFinder uses a model-based approach to calculate gene stability values based on both inter- and intra- group expression variation (Andersen et al., 2004). NormFinder also suggests the best pair of genes and calculates a stability value for this combination of genes. We divided our sample sets into groups based on sex (brain and pituitary, both species), plumage morph (sparrow) or breeding stage (sparrow) for the NormFinder analysis.
For both geNorm and NormFinder analyses, Cp values were converted to relative quantities. Finch Cp values were converted to relative quantities using a standard curve run on the same plate as the samples. Because space constraints prevented us from running standard curves on the same plate as the sparrow samples, all sparrow Cp values were transformed to relative quantities with the highest relative quantity set to one using the equation 2−ΔCp where ΔCP = Cpsample − Cpmin.
To test whether the choice of reference gene might affect the outcome of a study, we evaluated the expression of example target genes in zebra finch brain, sparrow pituitary, and sparrow testis. In zebra finch brain, we measured the expression of apolipoprotein D (APOD), a gene that was previously shown to be expressed at higher levels in males than females (Naurin et al., 2011). We normalized APOD expression to the best combination of reference genes (Tables 3 and 4) as determined by both geNorm and NormFinder (GAPDH and PPIA). In addition, to test the extent to which reference gene choice affects sensitivity to detect an effect, we normalized APOD expression to individual reference genes. Normalized expression values (target gene expression / reference gene expression) were calculated using LightCycler 480 software (Roche, Advanced relative quantification).
Table 3.
Reference genes are listed from most to least stable, according to geNorm analysis, in zebra finch and white-throated sparrow brain, pituitary, ovary, and testis. The average expression stability value (M) is listed next to each gene.a
| Zebra finch | White-throated Sparrow |
|||
|---|---|---|---|---|
| Tissue | Gene | M | Gene | M |
| Brain | GAPDH | 0.108 | RPL4 | 0.276 |
| PPIA | 0.108 | PPIA | 0.276 | |
| RPL4 | 0.127 | TFRC | 0.403 | |
| RPL32 | 0.133 | HPRT | 0.488 | |
| ACTB | 0.145 | GAPDH | 0.567 | |
| HPRT | 0.156 | RPL32 | 0.612 | |
| 18S | 0.180 | 18S | 0.666 | |
| TFRC | 0.201 | |||
| UBC | 0.237 | |||
| Pituitary | RPL4 | 0.124 | 18S | 0.347 |
| RPL32 | 0.124 | GAPDH | 0.347 | |
| PPIA | 0.177 | HPRT | 0.387 | |
| GAPDH | 0.208 | ACTB | 0.485 | |
| 18S | 0.233 | HMBS | 0.621 | |
| HPRT | 0.253 | |||
| ACTB | 0.297 | |||
| UBC | 0.347 | |||
| TFRC | 0.392 | |||
| Ovary | ACTB | 0.115 | HMBS | 0.266 |
| PPIA | 0.115 | HPRT | 0.266 | |
| RPL4 | 0.197 | GAPDH | 0.288 | |
| RPL32 | 0.225 | PPIA | 0.311 | |
| GAPDH | 0.253 | RPL4 | 0.327 | |
| UBC | 0.301 | RPL32 | 0.364 | |
| 18S | 0.340 | ACTB | 0.402 | |
| HPRT | 0.402 | TFRC | 0.455 | |
| TFRC | 0.479 | 18S | 0.571 | |
| Testis | RPL4 | 0.235 | HPRT | 0.279 |
| RPL32 | 0.235 | PPIA | 0.279 | |
| PPIA | 0.242 | RPL4 | 0.305 | |
| ACTB | 0.257 | RPL32 | 0.340 | |
| HPRT | 0.287 | ACTB | 0.384 | |
| TFRC | 0.362 | GAPDH | 0.434 | |
| 18S | 0.446 | TFRC | 0.464 | |
| UBC | 0.544 | HMBS | 0.602 | |
| GAPDH | 0.609 | 18S | 0.753 | |
Because gene ratios are used for stability measurements, the two most stable genes cannot be ranked in order.
Table 4.
Expression stability values for zebra finch samples, calculated by NormFinder. Samples were grouped by sex and NormFinder was used to calculate expression stability values (M) for individual genes and the best combination of two genes. Genes are listed from most to least stable (lowest to highest M value).
| Tissue | Gene | M | |
|---|---|---|---|
| Brain | |||
| Best combination | GAPDH, PPIA | 0.013 | |
| Most stable overall | PPIA | 0.013 | |
| GAPDH | 0.022 | ||
| RPL32 | 0.024 | ||
| RPL4 | 0.024 | ||
| ACTB | 0.028 | ||
| HPRT | 0.032 | ||
| 18S | 0.050 | ||
| TFRC | 0.051 | ||
| UBC | 0.075 | ||
| Pituitary | |||
| Best combination | PPIA, RPL4 | 0.017 | |
| Most stable overall | PPIA | 0.023 | |
| RPL4 | 0.024 | ||
| RPL32 | 0.030 | ||
| GAPDH | 0.042 | ||
| 18S | 0.059 | ||
| HPRT | 0.065 | ||
| ACTB | 0.082 | ||
| UBC | 0.117 | ||
| TFRC | 0.120 | ||
White-throated sparrows occur in two plumage morphs, white-striped and tan-striped, that differ from each other with respect to the number of GnRH neurons, plasma levels of luteinizing hormone, and sex steroids (Lake et al., 2008; Maney, 2008; Spinney et al., 2006). These differences are ultimately caused by a chromosomal rearrangement, present in white-striped but not tan-striped birds, that has captured a number of HPG-related genes. Here, we quantified the expression of two such genes, chorionic gonadotropin alpha polypeptide (CGA; the alpha chain of luteinizing hormone) and follicle-stimulating hormone receptor (FSHR). Our rationale for choosing these two genes was as follows: First, both play key roles in HPG regulation and both have been captured by the rearrangement. We measured expression of CGA in pituitaries collected from females because we previously showed that plasma levels of luteinizing hormone differ between morphs in females (Lake et al., 2008). We measured expression of FSHR in males early during the breeding season because the expression of this receptor is directly related to the rate of testis growth (Farner et al., 1981; Ishii & Farner, 1976) and white-striped males have larger testes during this breeding stage than tan-striped males (Horton et al., in press). As was done for the zebra finch analysis, we first normalized target gene expression to the top pair of reference genes identified by geNorm (Table 3), or to the top pair identified by the NormFinder analysis of morph (Table 5). We also report here the results of normalization to individual reference genes. Normalized expression values were calculated as above. Because not all of the white-throated sparrow samples could be run on the same plate, an arbitrary cDNA sample was included on every plate and used as a calibrator.
Table 5.
Expression stability values (M) for white-throated sparrow samples, as calculated by NormFinder. M values for individual genes and the best combination of two genes in brain, pituitary, ovary, or testis samples were calculated after grouping by plumage morph, breeding stage, or sex.a
| Morph | Stage | Sex | |||||
|---|---|---|---|---|---|---|---|
| Tissue | Gene | M | Gene | M | Gene | M | |
| Brain | |||||||
| Best combination | PPIA, TFRC | 0.023 | PPIA, TFRC | 0.023 | RPL4, TFRC | 0.039 | |
| Most stable overall | TFRC | 0.027 | TFRC | 0.028 | TFRC | 0.044 | |
| PPIA | 0.037 | PPIA | 0.036 | RPL4 | 0.072 | ||
| GAPDH | 0.044 | GAPDH | 0.042 | GAPDH | 0.075 | ||
| RPL4 | 0.050 | RPL4 | 0.050 | PPIA | 0.076 | ||
| RPL32 | 0.061 | RPL32 | 0.059 | 18S | 0.091 | ||
| HPRT | 0.071 | HPRT | 0.070 | HPRT | 0.117 | ||
| 18S | 0.075 | 18S | 0.076 | RPL32 | 0.121 | ||
| Pituitary | |||||||
| Best combination | 18S, HPRT | 0.029 | 18S, GAPDH | 0.018 | ACTB, GAPDH | 0.041 | |
| Most stable overall | GAPDH | 0.035 | GAPDH | 0.017 | HPRT | 0.053 | |
| 18S | 0.036 | 18S | 0.032 | GAPDH | 0.067 | ||
| HPRT | 0.043 | HPRT | 0.039 | 18S | 0.067 | ||
| ACTB | 0.062 | ACTB | 0.059 | HMBS | 0.090 | ||
| HMBS | 0.082 | HMBS | 0.084 | ACTB | 0.108 | ||
| Ovary | |||||||
| Best combination | HMBS, PPIA | 0.018 | HMBS, PPIA | 0.022 | |||
| Most stable overall | HMBS | 0.022 | HMBS | 0.022 | |||
| PPIA | 0.029 | PPIA | 0.028 | ||||
| GAPDH | 0.030 | GAPDH | 0.030 | ||||
| RPL4 | 0.037 | RPL4 | 0.041 | ||||
| HPRT | 0.044 | HPRT | 0.043 | ||||
| ACTB | 0.062 | RPL32 | 0.059 | ||||
| RPL32 | 0.065 | ACTB | 0.074 | ||||
| TFRC | 0.111 | TFRC | 0.115 | ||||
| 18S | 0.168 | 18S | 0.172 | ||||
| Testis | |||||||
| Best combination | HPRT, PPIA | 0.032 | HPRT, PPIA | 0.027 | |||
| Most stable overall | HPRT | 0.037 | HPRT | 0.055 | |||
| PPIA | 0.041 | RPL4 | 0.057 | ||||
| RPL4 | 0.057 | RPL32 | 0.065 | ||||
| RPL32 | 0.068 | PPIA | 0.066 | ||||
| GAPDH | 0.082 | ACTB | 0.099 | ||||
| TFRC | 0.097 | TFRC | 0.149 | ||||
| ACTB | 0.101 | GAPDH | 0.153 | ||||
| HMBS | 0.149 | HMBS | 0.170 | ||||
| 18S | 0.185 | 18S | 0.204 | ||||
Although the rankings changed slightly according to whether the samples were grouped by sex, morph, or stage, the overall patterns were similar. In brain, ranks 2-4 were occupied by PPIA, GAPDH, and RPL4 in all three analyses. In pituitary, GAPDH, 18S, and HPRT were always the top three genes. In ovary, the top five genes were the same regardless of group. In testis, the top four genes were HPRT, PPIA, RPL4, and RPL32. To determine the best combination of two genes, Normfinder uses intergroup stability calculations. Thus, although the top two genes were most often the best combination, other best combinations were sometimes identified, as was the case in pituitary (by sex) and testis (by stage).
Statistical analysis of target gene expression
We used t tests to compare the expression of target genes between groups. Effect size was calculated using Cohen’s d. Because our analysis of CGA in pituitary contained samples from both breeding stages (pre-parental and parental), we compared across morph while controlling for stage using 2-way ANOVA with morph and stage as fixed factors (SPSS Inc, Chicago IL). SPSS software was also used to calculate partial Eta squared (effect size) and estimated marginal means ± SEM.
Results
We evaluated the stability of reference gene expression using two computer programs: geNorm and NormFinder. We used geNorm to rank genes from most to least stable in each set of samples. The geNorm program calculated the pair of genes with the lowest average expression stability value, and ranked the rest of the genes according to M values (Table 3). Many of the genes in Table 3 showed average expression stability values <0.5, indicating that these genes were stably expressed in our samples (Hellemans et al., 2007). The optimal number of control genes required for accurate normalization was determined by pairwise variation analysis (Vandesompele et al., 2002). For each set of samples, normalization using the two most stable reference genes was sufficient, as the addition of the third most stable gene did not make a significant contribution to the normalization factor (Supp. Fig. 1).
We used NormFinder to assess the stability of reference genes across sex for the zebra finches, and across sex, morph, and breeding stage for the sparrows. NormFinder calculated M values for each individual gene and for the best combination of two genes across groups (Tables 4 and 5). Although the optimal combination of two genes suggested by NormFinder varied slightly according to sample grouping, the top genes in each sample set were consistently stably expressed, even when compared across sex, breeding stage, and plumage morph. NormFinder was generally in agreement with geNorm regarding the top-ranked genes.
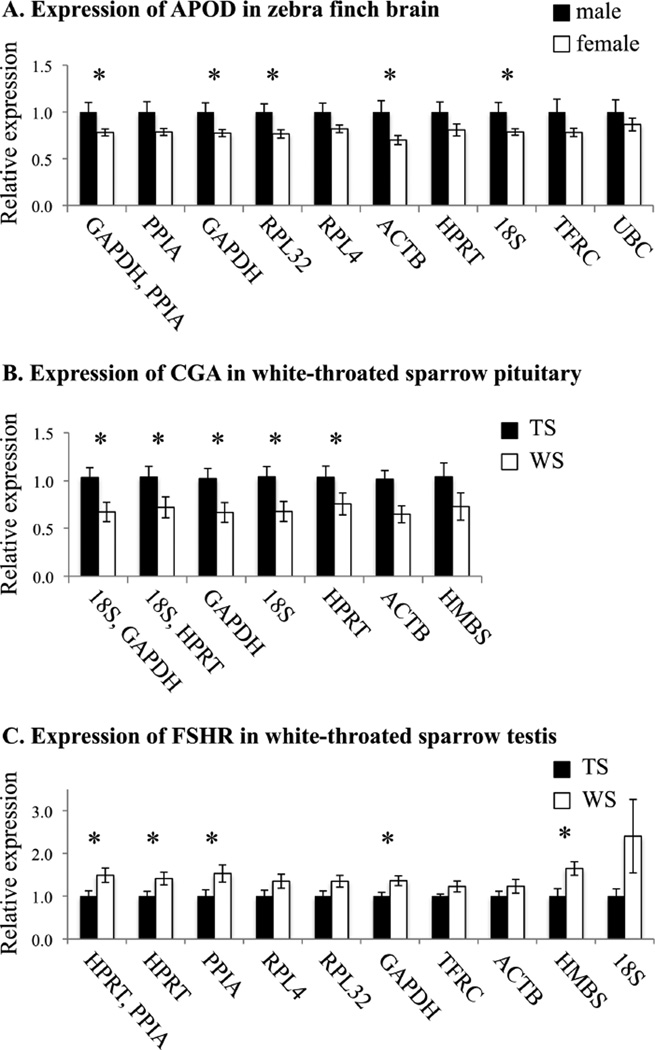
In order to assess the extent to which reference gene choices may affect experimental outcome, we used our results to quantify the relative expression of example target genes in both zebra finch and sparrow tissues. We first measured expression of APOD, a gene known to be more highly expressed in the brains of male than female zebra finches (Naurin et al., 2011). A sex difference in expression was detected when expression was normalized to GAPDH and PPIA, the combination recommended by both geNorm and NormFinder for a comparison across sex (p=0.042; Fig. 1A and Table 6). We also detected a sex difference when using GAPDH, RPL32, ACTB, and 18S individually, reflecting the high stability of many of the selected reference genes. When we normalized APOD expression to the worst performing reference genes, however, the difference in expression between males and females was lost, resulting in a type II statistical error.
Figure 1.
Target gene expression when normalized to the reference genes evaluated in this study. A) Expression of APOD in zebra finch brain. B) Expression of CGA in female white-throated sparrow pituitary. C) Expression of FSHR in white-throated sparrow testis. Reference genes used for normalization are indicated on the x-axis, in the order recommended by the software program NormFinder (see text). In order to plot all of the ratios on the same graph, all APOD ratios were normalized to the average male value and all CGA and FSHR ratios were normalized to the average tanstriped value. Values for CGA are represented as estimated marginal means. Error bars represent standard error of the mean (A,C) or standard error of the estimated marginal mean (B). APOD: Apolipoprotein D; CGA: chorionic gonadotropin alpha polypeptide; FSHR: follicle stimulating hormone receptor; TS: tanstriped; WS: white-striped. *p value < 0.05; see Table 6 for statistics.
Table 6.
Detection of intergroup differences in target gene expression using the reference genes evaluated in this study.a
| Reference Gene(s) | p-value | Effect size |
|---|---|---|
| Sex difference in APOD expression in zebra finch brain | ||
| GAPDH, PPIA | 0.042 | 0.944 |
| PPIA | 0.062 | 0.856 |
| GAPDH | 0.031 | 1.00 |
| RPL32 | 0.020 | 1.12 |
| RPL4 | 0.075 | 0.818 |
| ACTB | 0.023 | 1.08 |
| HPRT | 0.124 | 0.710 |
| 18S | 0.044 | 0.934 |
| TFRC | 0.117 | 0.708 |
| UBC | 0.349 | 0.420 |
| Morph difference in CGA expression in white-throated sparrow pituitary | ||
| 18S, GAPDH | 0.030 | 0.164 |
| 18S, HPRT | 0.042 | 0.163 |
| GAPDH | 0.024 | 0.145 |
| 18S | 0.036 | 0.152 |
| HPRT | 0.040 | 0.152 |
| ACTB | 0.083 | 0.107 |
| HMBS | 0.114 | 0.090 |
| Morph difference in FSHR expression in white-throated sparrow testis | ||
| HPRT, PPIA | 0.030 | 0.930 |
| HPRT | 0.039 | 0.880 |
| PPIA | 0.047 | 0.845 |
| RPL4 | 0.119 | 0.651 |
| RPL32 | 0.071 | 0.760 |
| GAPDH | 0.022 | 0.991 |
| TFRC | 0.122 | 0.654 |
| ACTB | 0.261 | 0.465 |
| HMBS | 0.012 | 1.10 |
| 18S | 0.138 | 0.628 |
Target genes apolipoprotein D (APOD), chorionic gonadotropin alpha polypeptide (CGA), and follicle stimulating hormone receptor (FSHR) were normalized to the reference genes indicated in the first column. Samples were grouped by sex (APOD) or morph (CGA and FSHR) for calculations of p-value and effect size (Cohen’s d for APOD and FSHR; Eta squared for CGA). p-values <0.05 are in bold.
In white-throated sparrows, we measured the expression of two genes that we expected to differ between the morphs because they have been captured by the chromosomal rearrangement linked to morph (Thorneycroft, 1981; Thomas et al., 2008) and because our previous research led us to suspect morph differences in these genes (e.g. Lake et al., 2008). When CGA was normalized to the best pair of genes as determined by either geNorm or NormFinder for comparison across morph, a significant difference in expression was detected (p= 0.030 and 0.042 respectively; Fig. 1B and Table 6). CGA was expressed at higher levels in the pituitaries of tan-striped than in white-striped females. This morph difference was also detectable when CGA expression was normalized to GAPDH, 18S, or HPRT individually, but not when normalized to the lower-ranked genes ACTB or HMBS.
FSHR was more highly expressed in the testes of white-striped than tan-striped males. This difference in expression was detected when FSHR expression was normalized to HPRT and PPIA, HPRT, PPIA, GAPDH, or HMBS (Fig. 1C and Table 6). No morph difference was detected when expression was normalized to RPL4, RPL32, TFRC, ACTB, or 18S. The greatest difference between morphs was detected when FSHR was normalized to HMBS (p= 0.012 and effect size 1.10, Table 6). To better understand why one of the lowest-ranked genes might have produced the largest effect size, we used NormFinder to calculate intergroup variation for these samples. Large intergroup variation can lead to over- or underestimating actual differences between two groups (Andersen et al., 2004). HMBS had the largest positive intergroup variation of all genes tested: 0.232, vs. −0.015 and 0.023 for HPRT and PPIA, respectively. This positive value indicates that HMBS showed systematically higher expression in the tanstriped samples, which would lead to overestimating a morph difference when the target gene is more highly expressed in white-striped birds-- as is the case with FSHR. Conversely, TFRC and ACTB had large negative intergroup variation values (−0.127 and −0.118), which likely led to underestimating a morph difference.
Discussion
Songbird models are rich resources for understanding the neuroendocrine basis of complex social behavior. Emerging technologies allowing easy and rapid quantification of neuroendocrine gene expression, in a variety of songbird species and tissues, could thus have great impact on our understanding of underlying mechanisms. Reference genes for qPCR have been validated for a number of tissues in multiple organisms (Boda et al., 2009; Cheng et al., 2009; De Boever et al., 2008; de Jonge et al., 2007; Vandesompele et al., 2002; Yin et al., 2010; Yue et al., 2010), but to date there is little information on appropriate reference genes in tissues of songbirds. In a survey of studies using qPCR to measure gene expression in songbird neural and endocrine tissue, we found that the most common reference genes were 18S, ACTB, and GAPDH (Table 1). Despite their popularity, however, these genes are not always appropriate for normalization (Bentley et al., 2013b; Lombardino et al., 2006; Perfito et al., 2012; Wada et al., 2006). Thus, there is a need to identify additional genes that may serve as suitable choices.
In this study, we evaluated reference genes for use in brain, pituitary, and gonad in two common songbird models. The results of our analysis highlight several important points to consider. First, even the genes identified as top-performers by NormFinder or geNorm may not be adequate when used alone. For example, we were unable to detect a significant sex difference in APOD in zebra finch brain when using the top-ranked NormFinder gene PPIA (Fig 1A). When we used the recommended combination of two genes, however, the difference was significant. Like other authors have done, we recommend that a combination of validated reference genes be used when possible (Bustin et al., 2009; Vandesompele et al., 2002).
Second, the stability of reference genes varies according to species. For example TFRC, which in sparrow brain was ranked highly by NormFinder (Table 4), was much less stably expressed in zebra finch brain. When we used TFRC to normalize samples in an example comparison in zebra finches, we could not detect an effect that was detectable using other genes (Fig 1A). Thus, although our tables provide a good starting point with which to choose genes, it is quite important to validate these genes for species other than the ones used here. Similarly, the expression of these genes varies according to tissue. geNorm ranked 18S as the most stable gene in sparrow pituitary, but the least stable in the brain, ovary, and testis of the same species. This difference is exemplified in our analysis of target gene expression in sparrow tissues. When we normalized CGA, an example target gene, to 18S in pituitary, we were able to detect a difference in expression between morphs. When FSHR expression was compared between morphs in testis, a difference in expression detected when normalizing to the top reference genes was lost when normalizing to 18S (Fig 1C). Researchers using tissues other than the ones we used in this study should therefore be mindful that our results may not extrapolate to other tissues.
Third, we emphasize that reference genes must be stable across experimental groups and conditions. Here, we tested the stability of genes that could be used to detect effects of sex in zebra finches, and effects of sex, morph, and breeding stage (pre-parental vs. parental) in white-throated sparrows. The most stable genes in our analyses appeared to retain stability across a variety of conditions (sex, morph, and stage, Table 5). Intergroup variation can lead to type I or type II statistical error, depending on the direction of bias (Andersen et al., 2004). Indeed, we found that HMBS, TFRC, and ACTB appeared to differ between morphs in sparrow testis. Although we found that normalization to HMBS led to only a slight overestimation of effect size (Table 6), large effects of the variable of interest on reference gene expression could result in erroneous conclusions. In our study, we did not test for stability across common experimental conditions such as daylength, hormonal state, or exposure to conspecific signals. The reference genes used in any study should be evaluated, for example using NormFinder or geNorm, for stability under the experimental conditions specific to that study (Bustin et al., 2009).
Finally, in addition to stability, relative expression level should be considered when selecting reference genes. Despite its ranking by both programs as one of the most stable genes in sparrow ovary (Tables 3 and 4), HMBS was poorly expressed and therefore is not recommended as a reference gene in this tissue (data not shown).
Overall, although our analysis cannot replace the need for validation in other model organisms, tissues, and experimental conditions, our results are consistent with the idea that reference genes shown to be suitable in a variety of species and tissues may perform well in species and tissues not yet tested. Indeed, our list of candidate reference genes consisted of those that are stably expressed in multiple tissues and organisms; we show here that many of these genes are also stable in songbird tissues (geNorm average M values <0.5). We hope that our data will help other researchers avoid making poor choices and to select reference genes for further validation in other songbird studies.
Supplementary Material
Highlights.
We evaluated ten reference genes for quantitative real-time-PCR in songbirds
Reference genes were evaluated in brain, pituitary and gonad of two
18S, GAPDH, ACTB were not always the best choice of reference gene
We identified reference genes more suitable for use in these tissues
Acknowledgements
We are grateful to Laura Carruth for providing zebra finches, and to the Emory University Departments of Biology and Human Genetics for use of resources. We thank Jake Metzler and the Forest Society of Maine for access to our field site, the Hemlock Stream Forest. We are obliged to the University of Maine School of Biology and Ecology and Clarissa Henry for logistical support in Maine. We thank Demesew Abebe for technical assistance and Adela Annis, Elaina Burns, Jenna Cava, Allison Cornell, Chris Gurguis, Clifton McKee, and Justin Michaud for field assistance. This work was supported by NIMH 1R01MH082833-01A2 to DLM and NSF SMA-1306132 to WZK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References Cited
- Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Banerjee SB, Arterbery AS, Fergus DJ, Adkins-Regan E. Deprivation of maternal care has long-lasting consequences for the hypothalamic-pituitary-adrenal axis of zebra finches. Proc. Biol. Sci. 2012;279:759–766. doi: 10.1098/rspb.2011.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley GE, Perfito N, Calisi RM. Season- and context-dependent sex differences in melatonin receptor activity in a forebrain song control nucleus. Horm. Behav. 2013a;63:829–835. doi: 10.1016/j.yhbeh.2012.11.015. [DOI] [PubMed] [Google Scholar]
- Bentley GE, Tucker S, Chou H, Hau M, Perfito N. Testicular growth and regression are not correlated with Dio2 expression in a wild male songbird, sturnus vulgaris, exposed to natural changes in photoperiod. Endocrinology. 2013b;154:1813–1819. doi: 10.1210/en.2013-1093. [DOI] [PubMed] [Google Scholar]
- Bergeon Burns CM, Rosvall KA, Hahn TP, Demas GE, Ketterson ED. Examining sources of variation in HPG axis function among individuals and populations of the dark-eyed junco. Horm. Behav. 2014;65:179–187. doi: 10.1016/j.yhbeh.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeon Burns CM, Rosvall KA, Ketterson ED. Neural steroid sensitivity and aggression: comparing individuals of two songbird subspecies. J. Evol. Biol. 2013;26:820–831. doi: 10.1111/jeb.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boda E, Pini A, Hoxha E, Parolisi R, Tempia F. Selection of reference genes for quantitative real-time RT-PCR studies in mouse brain. J. Mol. Neurosci. 2009;37:238–253. doi: 10.1007/s12031-008-9128-9. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: Minimun information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Cerasale DJ, Zajac DM, Guglielmo CG. Behavioral and physiological effects of photoperiod-induced migratory state and leptin on a migratory bird, Zonotrichia albicollis: I. anorectic effects of leptin administration. Gen. Comp. Endocrinol. 2011;174:276–286. doi: 10.1016/j.ygcen.2011.08.025. [DOI] [PubMed] [Google Scholar]
- Cheng WC, Chang CW, Chen CR, Tsai ML, Shu WY, Li CY, Hsu IC. Identification of reference genes across physiological states for qRT-PCR through microarray meta-analysis. PloS One. 2011;6:e17347. doi: 10.1371/journal.pone.0017347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton DF, London SE. Advancing avian behavioral neuroendocrinology through genomics. Front. Neuroendocrinol. 2014;35:58–71. doi: 10.1016/j.yfrne.2013.09.004. [DOI] [PubMed] [Google Scholar]
- De Boever S, Vangestel C, De Backer P, Croubels S, Sys SU. Identification and validation of housekeeping genes as internal control for gene expression in an intravenous LPS inflammation model in chickens. Vet. Immunol. Immunopathol. 2008;122:312–317. doi: 10.1016/j.vetimm.2007.12.002. [DOI] [PubMed] [Google Scholar]
- de Jonge HJ, Fehrmann RS, de Bont ES, Hofstra RM, Gerbens F, Kamps WA, de Vries EG, van der Zee AG, te Meerman GJ, ter Elst A. Evidence based selection of housekeeping genes. PloS One. 2007;2:e898. doi: 10.1371/journal.pone.0000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan KA, Moon J, Vartosis D, Zee I. Injury-induced expression of glial androgen receptor in the zebra finch brain. J. Neurotrauma. 2013;30:1919–1924. doi: 10.1089/neu.2013.2951. [DOI] [PubMed] [Google Scholar]
- Duncan KA, Saldanha CJ. Neuroinflammation induces glial aromatase expression in the uninjured songbird brain. J. Neuroinflammation. 2011;8:81. doi: 10.1186/1742-2094-8-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egbert JR, Jackson MF, Rodgers BD, Schwabl H. Between-female variation in house sparrow yolk testosterone concentration is negatively associated with CYP19A1 (aromatase) mRNA expression in ovarian follicles. Gen. Comp. Endocrinol. 2013;183:53–62. doi: 10.1016/j.ygcen.2012.12.001. [DOI] [PubMed] [Google Scholar]
- Farner DS, Donham RS, Moore MC. Induction of testicular development in house sparrows, Passer domesticus, and white-crowned sparrows, Zonotrichia leucophrys gambelii, with very long days and continuous light. Physiol. Zool. 1981;54:372–378. [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biology. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton BM, Hu Y, Martin CL, Bunke BP, Matthews BS, Moore IT, Thomas JW, Maney DL. Behavioral characterization of a white-throated sparrow homozygous for the ZAL2(m) chromosomal rearrangement. Behav. Genet. 2013;43:60–70. doi: 10.1007/s10519-012-9574-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton BM, Hudson WH, Ortland EA, Shirk S, Thomas JW, Young ER, Zinzow-Kramer WM, Maney DL. Estrogen receptor a polymorphism in a species with alternative behavioral phenotypes. Proc. Natl. Acad. Sci. U.S.A. 2014;111:1443–1448. doi: 10.1073/pnas.1317165111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton BM, Moore IT, Maney DM. New Insights into the Hormonal and Behavioural Correlates of Polymorphism in White-throated Sparrows. Anim. Behav. doi: 10.1016/j.anbehav.2014.04.015. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel TR, Fernald RD. How the brain processes social information: searching for the social brain. Annu. Rev. Neurosci. 2004;27:697–722. doi: 10.1146/annurev.neuro.27.070203.144148. [DOI] [PubMed] [Google Scholar]
- Ishii S, Farner DS. Binding of follicle-stimulating hormone by homogenates of testes of photostimulated white-crowned sparrows, Zonotrichia leucophrys gambelii. Gen. Comp. Endocrinol. 1976;30:443–450. doi: 10.1016/0016-6480(76)90113-1. [DOI] [PubMed] [Google Scholar]
- Jasnow AM, Schulkin J, Pfaff DW. Estrogen facilitates fear conditioning and increases corticotropin-releasing hormone mRNA expression in the central amygdala in female mice. Horm. Behav. 2006;49:197–205. doi: 10.1016/j.yhbeh.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Jones S, Pfister-Genskow M, Benca RM, Cirelli C. Molecular correlates of sleep and wakefulness in the brain of the white-crowned sparrow. J. Neurochem. 2008a;105:46–62. doi: 10.1111/j.1471-4159.2007.05089.x. [DOI] [PubMed] [Google Scholar]
- Jones S, Pfister-Genskow M, Cirelli C, Benca RM. Changes in brain gene expression during migration in the white-crowned sparrow. Brain Res. Bull. 2008b;76:536–544. doi: 10.1016/j.brainresbull.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Lilliehook C, Roides B, Chen Z, Chang M, Mobashery S, Goldman SA. Testosterone-induced matrix metalloproteinase activation is a checkpoint for neuronal addition to the adult songbird brain. J. Neurosci. 2008;28:208–216. doi: 10.1523/JNEUROSCI.3674-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake JI, Lange HS, O’Brien S, Sanford SE, Maney DL. Activity of the hypothalamic-pituitary-gonadal axis differs between behavioral phenotypes in female white-throated sparrows (Zonotrichia albicollis) Gen. Comp. Endocrinol. 2008;156:426–433. doi: 10.1016/j.ygcen.2007.12.009. [DOI] [PubMed] [Google Scholar]
- Levin BE, Dunn-Meynell AA, Banks WA. Obesity-prone rats have normal blood-brain barrier transport but defective central leptin signaling before obesity onset. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2004;286:R143–R150. doi: 10.1152/ajpregu.00393.2003. [DOI] [PubMed] [Google Scholar]
- Liebl AL, Martin LB. Stress hormone receptors change as range expansion progresses in house sparrows. Biol. Lett. 2013;9:20130181. doi: 10.1098/rsbl.2013.0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebl AL, Shimizu T, Martin LB. Covariation among glucocorticoid regulatory elements varies seasonally in house sparrows. Gen. Comp. Endocrinol. 2013;183:32–37. doi: 10.1016/j.ygcen.2012.11.021. [DOI] [PubMed] [Google Scholar]
- Lombardino AJ, Hertel M, Li XC, Haripal B, Martin-Harris L, Pariser E, Nottebohm F. Expression profiling of intermingled long-range projection neurons harvested by laser capture microdissection. J. Neurosci. Methods. 2006;157:195–207. doi: 10.1016/j.jneumeth.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Maney DL. Endocrine and genomic architecture of life history trade-offs in an avian model of social behavior. Gen. Comp. Endocrinol. 2008;157:275–282. doi: 10.1016/j.ygcen.2008.03.023. [DOI] [PubMed] [Google Scholar]
- Medina CO, Lattin CR, McVey M, Romero LM. There is no correlation between glucocorticoid receptor mRNA expression and protein binding in the brains of house sparrows (Passer domesticus) Gen. Comp. Endocrinol. 2013;193:27–36. doi: 10.1016/j.ygcen.2013.07.008. [DOI] [PubMed] [Google Scholar]
- Michopoulos V, Maney DL, Morehouse CB, Thomas JW, Rohwer S. A genotyping assay to determine plumage morph in the white-throated sparrow (Zonotrichia albicollis) The Auk. 2007;124:1330–1335. [Google Scholar]
- Mirzatoni A, Spence RD, Naranjo KC, Saldanha CJ, Schlinger BA. Injury-induced regulation of steroidogenic gene expression in the cerebellum. J. of Neurotrauma. 2010;27:1875–1882. doi: 10.1089/neu.2010.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai M, Replogle K, Drnevich J, Wang G, Wacker D, Band M, Clayton DF, Wingfield JC. Seasonal differences of gene expression profiles in song sparrow (Melospiza melodia) hypothalamus in relation to territorial aggression. PLoS One. 2009;4:e8182. doi: 10.1371/journal.pone.0008182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naurin S, Hansson B, Hasselquist D, Kim Y, Bensch S. The sex-biased brain: sexual dimorphism in gene expression in two species of songbirds. BMC Genomics. 2011;12:37. doi: 10.1186/1471-2164-12-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfito N, Jeong SY, Silverin B, Calisi RM, Bentley GE, Hau M. Anticipating spring: wild populations of great tits (Parus major) differ in expression of key genes for photoperiodic time measurement. PloS One. 2012;7:e34997. doi: 10.1371/journal.pone.0034997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LM, Mohr M, Wade J. Enhanced expression of tubulin-specific chaperone protein A, mitochondrial ribosomal protein S27, and the DNA excision repair protein XPACCH in the song system of juvenile male zebra finches. Dev. Neurobiol. 2012;72:199–207. doi: 10.1002/dneu.20956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Replogle K, Arnold AP, Ball GF, Band M, Bensch S, Brenowitz EA, Dong S, Drnevich J, Ferris M, George JM, Gong G, Hasselquist D, Kin R, Lewin HA, Liu L, Lovell PV, Mello CV, Naurin S, Rodriguez-Zas S, Thimmapuram J, Clayton DF. The Songbird Neurogenomics (SoNG) Initiative: community-based tools and strategies for study of brain gene function and evolution. BMC Genomics. 2008;9:131. doi: 10.1186/1471-2164-9-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson GE, Fernald RD, Clayton DF. Genes and Social Behavior. Science. 2008;322:896–899. doi: 10.1126/science.1159277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson GE, Grozinger CM, Whitfield CW. Sociogenomics: social life in molecular terms. Nat. Rev. Genet. 2005;6:257–270. doi: 10.1038/nrg1575. [DOI] [PubMed] [Google Scholar]
- Rosvall KA, Bergeon Burns CM, Barske J, Goodson JL, Schlinger BA, Sengelaub DR, Ketterson ED. Neural sensitivity to sex steroids predicts individual differences in aggression: implications for behavioural evolution. Proc. Biol. Sci. 2012;279:3547–3555. doi: 10.1098/rspb.2012.0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosvall KA, Bergeon Burns CM, Hahn TP, Ketterson ED. Sources of variation in HPG axis reactivity and individually consistent elevation of sex steroids in a female songbird. Gen. Comp. Endocrinol. 2013;194:230–239. doi: 10.1016/j.ygcen.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Luo G, Fu L, Fang Z, Wang X, Li X. miR-9 and miR-140-5p target FoxP2 and are regulated as a function of the social context of singing behavior in zebra finches. J. Neurosci. 2013;33:16510–16521. doi: 10.1523/JNEUROSCI.0838-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D, Rani S, Kumar V. Daily expression of six clock genes in central and peripheral tissues of a night-migratory songbird: Evidence for tissue-specific circadian timing. Chronobiol. Int. 2013;30:1208–1217. doi: 10.3109/07420528.2013.810632. [DOI] [PubMed] [Google Scholar]
- Spinney LH, Bentley GE, Hau M. Endocrine correlates of alternative phenotypes in the white-throated sparrow (Zonotrichia albicollis) Horm. Behav. 2006;50:762–771. doi: 10.1016/j.yhbeh.2006.06.034. [DOI] [PubMed] [Google Scholar]
- Stevenson TJ, Ball GF. Disruption of neuropsin mRNA expression via RNA interference facilitates the photoinduced increase in thyrotropin-stimulating subunit beta in birds. Eur, J. Neurosci. 2012;36:2859–2865. doi: 10.1111/j.1460-9568.2012.08209.x. [DOI] [PubMed] [Google Scholar]
- Thomas JW, Caceres M, Lowman JJ, Morehouse CB, Short ME, Baldwin EL, Maney DL, Martin CL. The chromosomal polymorphism linked to variation in social behavior in the white-throated sparrow (Zonotrichia albicollis) is a complex rearrangement and suppressor of recombination. Genetics. 2008;179:1455–1468. doi: 10.1534/genetics.108.088229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CK, Meitzen J, Replogle K, Drnevich J, Lent KL, Wissman AM, Farin F, Bammler TK, Beyer RP, Clayton DF, Perkel DJ, Brenowitz EA. Seasonal changes in patterns of gene expression in avian song control brain regions. PLoS One. 2012;7:e35119. doi: 10.1371/journal.pone.0035119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorneycroft HB. Chromosomal Polymorphism in the White-Throated Sparrow, Zonotrichia albicollis (Gmelin) Science. 1966;154:1571–1572. doi: 10.1126/science.154.3756.1571. [DOI] [PubMed] [Google Scholar]
- Throneycroft HB. A cytogenetic study of the White-Throated Sparrow, Zonotrichia albicollis (Gmelin) Evolution. 1975;29:611–621. doi: 10.1111/j.1558-5646.1975.tb00855.x. [DOI] [PubMed] [Google Scholar]
- Tomaszycki ML, Peabody C, Replogle K, Clayton DF, Tempelman RJ, Wade J. Sexual differentiation of the zebra finch song system: potential roles for sex chromosome genes. BMC Neurosci. 2009;10:24. doi: 10.1186/1471-2202-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanGuilder HD, Vrana KE, Freeman WM. Twenty-five years of quantitative PCR for gene expression analysis. BioTechniques. 2008;44:619–626. doi: 10.2144/000112776. [DOI] [PubMed] [Google Scholar]
- Wada K, Howard JT, McConnell P, Whitney O, Lints T, Rivas MV, Horita H, Patterson MA, White SA, Scharff C, Haesler S, Zhao S, Sakaguchi H, Hagiwara M, Shiraki T, Hirozane-Kishikawa T, Skene P, Hayashizaki Y, Carninci P, Jarvis ED. A molecular neuroethological approach for identifying and characterizing a cascade of behaviorally regulated genes. Proc. Natl. Acad. Sci. U.S.A. 2006;103:15212–15217. doi: 10.1073/pnas.0607098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade J, Tang YP, Peabody C, Tempelman RJ. Enhanced gene expression in the forebrain of hatchling and juvenile male zebra finches. J. Neurobiol. 2005;64:224–238. doi: 10.1002/neu.20141. [DOI] [PubMed] [Google Scholar]
- Warren WC, Clayton DF, Ellegren H, Arnold AP, Hillier LW, Kunstner A, Searle S, White S, Vilella AJ, Fairley S, Heger A, Kong L, Ponting CP, Jarvis ED, Mello CV, Minx P, Lovell P, Velho TA, Ferris M, Balakrishnan CN, Sinha S, Blatti C, London SE, Li Y, Lin YC, George J, Sweedler J, Southey B, Gunaratne P, Watson M, Nam K, Backstrom N, Smeds L, Nabholz B, Itoh Y, Whitney O, Pfenning AR, Howard J, Volker M, Skinner BM, Griffin DK, Ye L, McLaren WM, Flicek P, Quesada V, Velasco G, Lopez-Otin C, Puente XS, Olender T, Lancet D, Smit AF, Hubley R, Konkel MK, Walker JA, Batzer MA, Gu W, Pollock DD, Chen L, Cheng Z, Eichler EE, Stapley J, Slate J, Ekblom R, Birkhead T, Burke T, Burt D, Scharff C, Adam I, Richard H, Sultan M, Soldatov A, Lehrach H, Edwards SV, Yang SP, Li X, Graves T, Fulton L, Nelson J, Chinwalla A, Hou S, Mardis ER, Wilson RK. The genome of a songbird. Nature. 2010;464:757–762. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield JC, Farner DS. The endocrinology of wild species. In: Farner DS, King JR, Parkes KC, editors. Avian Biology. New York: Academic Press; 1993. pp. 163–327. [Google Scholar]
- Wynne RD, Maas S, Saldanha CJ. Molecular characterization of the injury-induced aromatase transcript in the adult zebra finch brain. J. Neurochem. 2008;105:1613–1624. doi: 10.1111/j.1471-4159.2008.05256.x. [DOI] [PubMed] [Google Scholar]
- Yin R, Tian F, Frankenberger B, de Angelis MH, Stoeger T. Selection and evaluation of stable housekeeping genes for gene expression normalization in carbon nanoparticle-induced acute pulmonary inflammation in mice. Biochem. Biophys. Res. Commun. 2010;399:531–536. doi: 10.1016/j.bbrc.2010.07.104. [DOI] [PubMed] [Google Scholar]
- Yue H, Lei XW, Yang FL, Li MY, Tang C. Reference gene selection for normalization of PCR analysis in chicken embryo fibroblast infected with H5N1 AIV. Virologica Sinica. 2010;25:425–431. doi: 10.1007/s12250-010-3114-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.