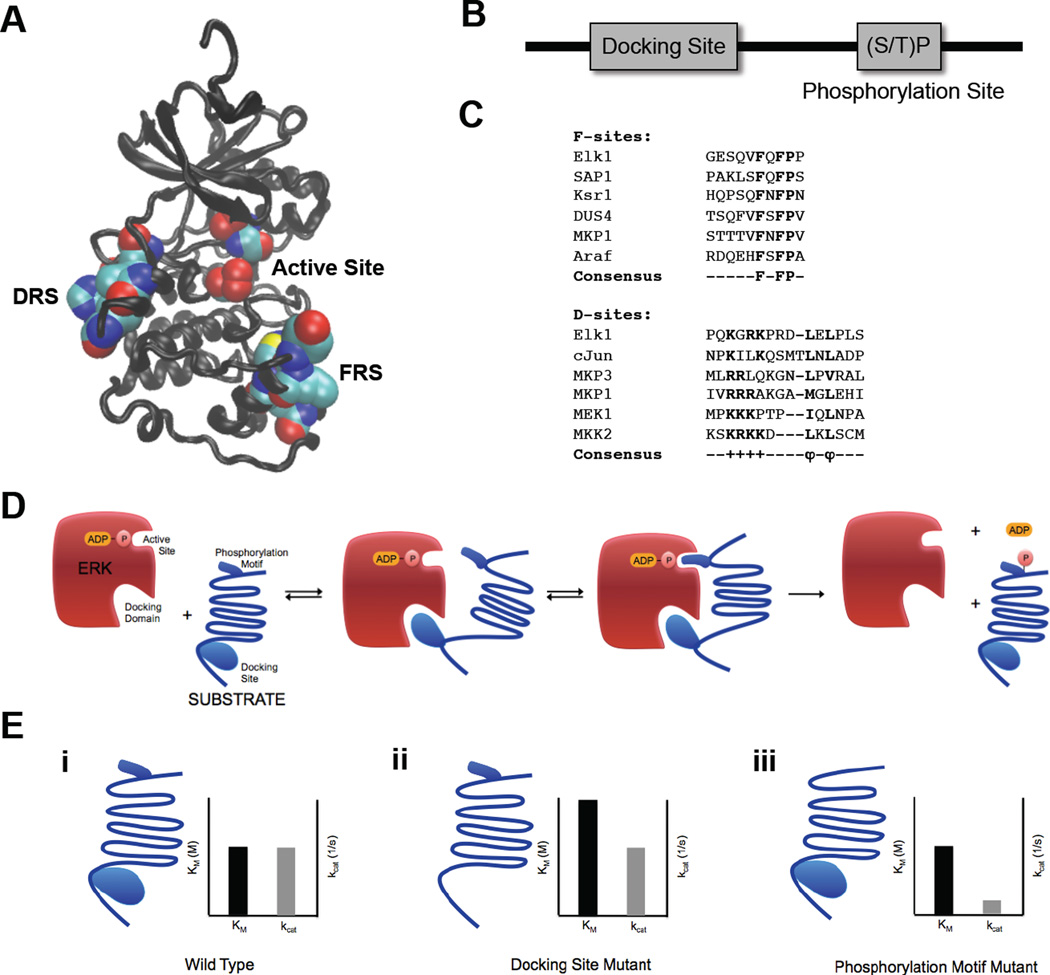
Figure 3.
ERK2 docking domains and ERK2 substrate docking sites. (A) ERK2 structure with docking domains – DRS domain (left) and FRS domain (right) – and active site (middle) in space filling representation. Structure drawn from PDB file 1ERK (33) (B) Schematic representation of ERK2 substrate sequence containing a docking site and a (S/T)P phosphorylation site. (C) Alignment of F-site and D-Site sequences from multiple ERK2 interacting substrates and regulators. (D) Schematic representation of ERK2 interactions with substrate docking site and (S/T)P phosphorylation motif. (E) Effects of mutations on substrate docking site and phosphorylation site and insights into molecular mechanisms of ERK2 catalysis. (i) Wild type substrate and associated Michaelis-Menten parameters. (ii) Substrate with docking site mutations and associated Michaelis-Menten parameters. Mutating the docking site significantly increases the Km but has little effect on kcat. (iii) Substrate with phosphorylation motif mutations and associated Michaelis-Menten parameters. Mutating the phosphorylation motif significantly decreases the kcat but has little effect on Km.