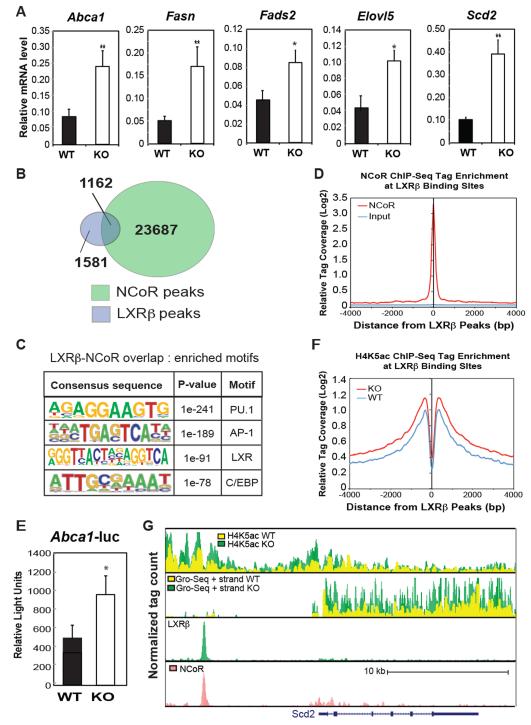
Figure 5.
Impact of NCoR KO on LXR target gene expression in macrophages. (A) Q-PCR analysis of the indicated LXR target genes in WT and MNKO macrophages. (B) Venn diagram of overlap between LXRβ and NCoR peaks in macrophages. (C) Sequence motifs associated with LXRβ and NCoR cobound sites. (D) Distribution of NCoR ChIP-Seq tag density in the vicinity of LXRβ binding sites. (E) LXR-dependent Abca1-luciferase reporter assays in WT and MNKO macrophages. (F) Distribution of H4K5ac ChIP-Seq tag density in the vicinity of LXRβ binding sites. (G) UCSC genome browser image illustrating normalized tag counts for H4K5Ac ChIP-Seq, GRO-Seq, LXRβ ChIP-Seq and NCoR ChIP-Seq (Barish et al., 2012) at the Scd2 locus. Values are expressed as mean ± s.e.m. * P<0.05, ** P<0.01 for KO versus WT.