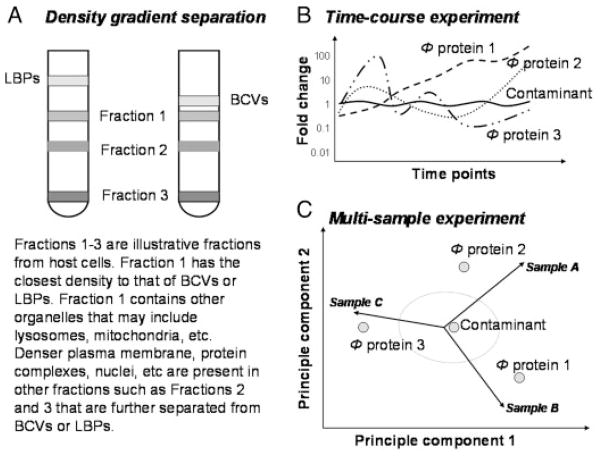
Figure 1.
Schematic illustration of phagosome preparation and a strategy to discern phagosome proteins from contaminating proteins during quantitative proteomic analyses of phagosomes. (A) In a sucrose density gradient separation of LBPs or BCVs from a host cell lysate, LBPs are better separated from other organelles due to their lighter density. (B) In a time-course experiment to monitor the changes in protein concentrations in a phagosome biogenesis, the concentration of a contaminant protein would not change over time because it does not participate in the biogenesis or maturation process of a phagosome. The proteins showing time-dependant fluctuation in concentration are most likely phagosome proteins. The figure was drawn based on [37, 39]. Four proteins are depicted. The proteins with dashed time-course curves are phagosome proteins and the one with a solid time-course curve represents a contaminating protein. Φ-phagosome. (C) In a multi-sample experiment where multiple phagosome preparations are compared under the same condition, the concentration of a contaminant protein would not differ among the several sample preparations. Proteins that are differentially regulated among the several phagosome preparations are most likely phagosome proteins. The figure was drawn based on [50], illustrating the biplot display of a principal component analysis of three phagosome preparations. Four proteins are depicted. The eclipse represents a confidence threshold.