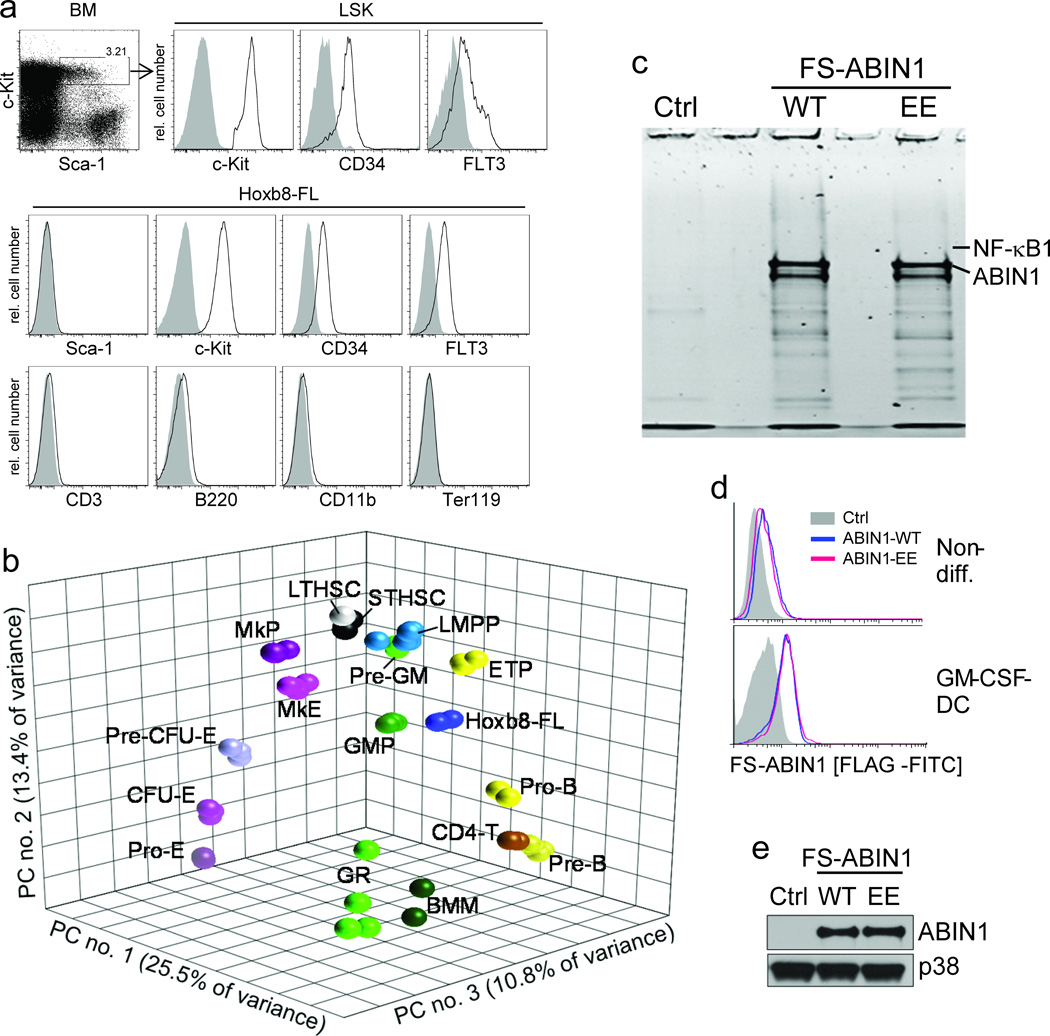
Figure 6. Phenotype, principal component analysis (PCA) and biochemical application of Hoxb8–FL cells.
(a) Lineage depleted BM cells and Hoxb8–FL cells were analyzed by flow cytometry. Open, specific antibodies; filled = isotype controls. Data are representative for at least five independent populations of Hoxb8–FL cells. (b) Three–dimensional PCA based on gene expression profiles of Hoxb8–FL cells (four independent populations), bone marrow–derived macrophages (BMM), granulocytes (GR) and purified hematopoietic progenitor cell populations. LTHSC, long–term hematopoietic stem cell; STHSC, short–term hematopoietic stem cell, LMPP, lymphoid–primed multipotent progenitor; ETP, early thymic progenitor; ProB, pro B–cell; PreB, pre B–cell; CD4T, CD4+ T–cell, Pre–GM, pre–granulocyte/ macrophage; GMP, granulocyte/macrophage progenitor; MkP, megakaryocyte progenitor; MkE, Megakaryocyte/ Erythrocyte progenitor; Pre CFU–E, pre–colony forming unit–erythrocytes; CFU–E, colony forming unit–erythrocytes; Pro–E, pro–Erythrocyte; GR, granulocyte. Each symbol represents an individual biological sample. PC, principal component. (c) Hoxb8–FL cells were generated from BM of ABIN1–deficient mice, reconstituted with epitope–tagged ABIN1 (FS–ABIN1), either wildtype (WT) or an ubiquitin–binding mutant (EE) or an empty control vector (Ctrl), differentiated into DC in the presence of GM–CSF for eight days, followed by tandem affinity purification of ABIN1. Purified proteins were separated by SDS/ PAGE, visualized by SYPRO Ruby (c) and identified by LC–MS (for peptide identifications see Supplementary Table 1). (d and e) ABIN1 expression was analyzed in non–differentiated cells (non–diff. (d)) and GM–CSF–driven DC (d, e) by intracellular (FLAG) staining and flow cytometry (d), and immuno–blotting with antibodies against ABIN1 (e).