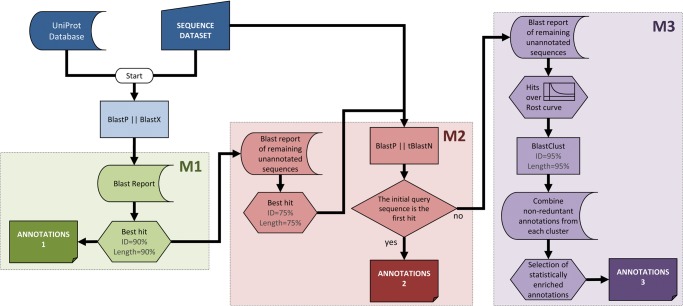
Figure 1.
Sma3s workflow. The first M1 module derives annotations from highly similar sequences stored in the database, choosing sequences using the Top-BLAST method, which selected the highest similarity homologue from each BLAST search that meets the minimum similarity criteria. The remaining sequences are passed to the second module (M2), which performs reciprocal BLAST searches to identify orthologous sequences as annotation sources, also using the Top-BLAST method. Finally, the M3 module obtains annotations from a set of related sequences whose similarity is supported by statistically significant concentrations of similar annotations, filtered by clustering techniques to avoid over-representation from duplicated gene families. This figure appears in colour in the online version of DNA Research.