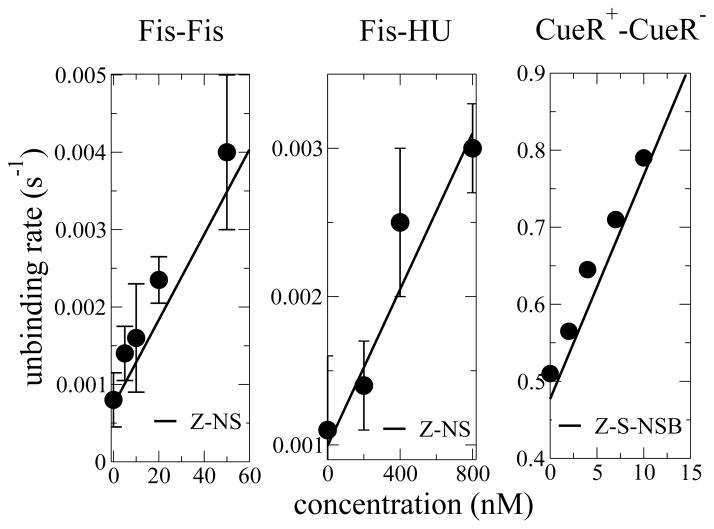
FIG. 3.
Fit of concentration-dependent unbinding rates of Fis bound to DNA in the presence of Fis (left) and HU (middle) proteins in solution [1], and for CueR dissociation rates as a function of CueR concentration in solution (right) [2], using a = 1 nm [8], and N=14, ε = 1.95 for Fis-Fis, N=19, ε = 1.4 for Fis-HU, and N=15, ε = 1.36 for CueR-CueR. For replacement rates (slopes of unbinding rates versus concentration) we find RFis−Fis = 5 104 M−1 s−1, RFis−HU = 2.6 103 M−1 s−1, and RCueR−CueR = 2.9 107 M−1 s−1, in agreement, considering the error bars, with experimental fits. Replacement concentrations found with the Z-NS model are cR = 2 nM for Fis- Fis and cR = 370 nM for Fis-HU, while we find cR = 16 nM for CueR-CueR with the Z-S-NSB model.