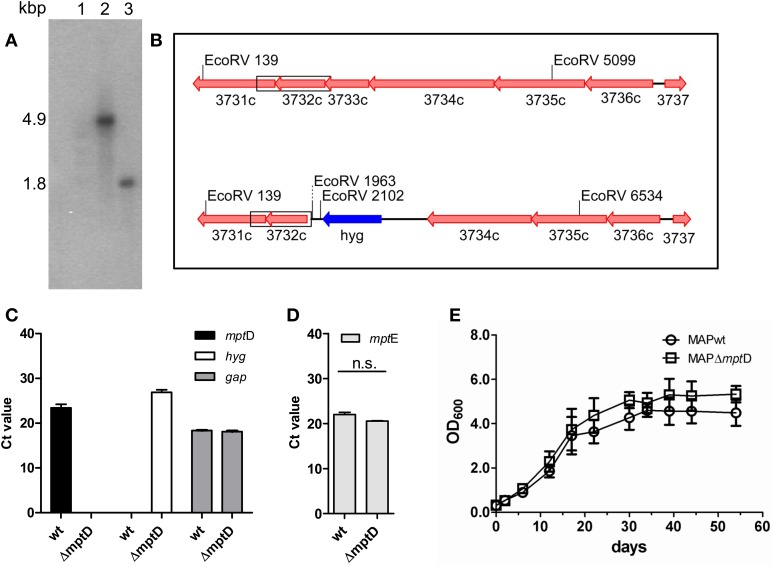
Figure 1.
Confirmation and characterization of MAPΔmptD mutant by Southern blot analysis (A,B), qRT-PCR (C,D) and growth experiments (E). (A) EcoRV restricted genomic DNA of MAPwt (lane 2) and MAPΔmptD mutant strain (lane 3), Southern blotted and hybridized with a probe against mptE (MAP3732c). In the wild type a 4.9 kbp fragment is labeled, the mutant strain shows a 1.8 kbp fragment. Lane 1 DNA marker. (B) Genomic sketch of the mpt region spanning genes mptF (MAP3731c) to mptA (MAP3736c) in MAPwt (top) and ΔmptD mutant strains (bottom). Position of EcoRV restriction sites are indicated, the α32-P-dCTP labeled probe fragment is boxed. (C) Transcription of mptD (black bars) and hygromycin (hyg) (white bars) in MAP wild type (wt) and MAPΔmptD were analyzed by qRT-PCR. As a control, cDNA samples were tested for the housekeeping gene gap (gray bars). (D) Analysis of mptE (MAP3732c) transcription in MAPwt and MAPΔmptD by qRT-PCR (n.s., no significant difference). (E) Growth of MAPwt and MAPΔmptD in Middlebrook 7H9 medium. Cultures were inoculated with an initial OD600 of 0.2 and growth was monitored at OD600 (ordinates) at different time points (abscissa) until stationary phase. The results of (C,D) represent the mean ± standard error (s.e.m.) of three replicates.