Fig. 3.
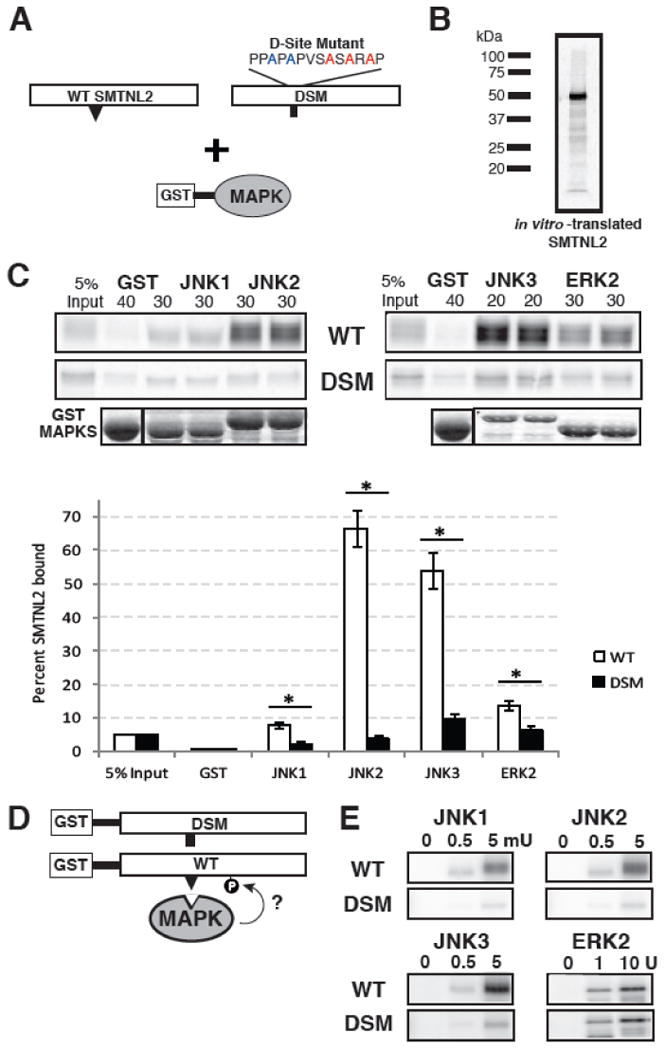
SMTNL2 is a novel substrate with a functional D-site. A) Full-length wild-type (WT) SMTNL2, or a D-site mutant (DSM) derivative, were tested for binding to GST-MAPK proteins. B) Representative in vitro transcription and translation product of full length SMTNL2. C) As shown in A, 35S-radiolabeled full-length SMTNL2 protein and a D-site mutant derivative were prepared by in vitro translation and partially purified by ammonium sulfate precipitation, and portions (5% of the amount added in the binding reactions) were resolved on a 10% SDS-polyacrylamide gel (lane 1 of each column of panels). Samples (∼1 pmol) of the same proteins were incubated with 40 μg of GST (lane 2 of each column) or with 30 μg of GST-JNK1, JNK2, JNK3 or ERK2 proteins (lanes 3-6 of each column, duplicate points shown), bound to glutathione-Sepharose beads, and the resulting bead-bound protein complexes were isolated by sedimentation, and resolved by 10% SDS-PAGE on the same gel. The gel was analyzed by staining with Coomassie Blue (CB) for visualization of the bound GST fusion protein (lowest row) and by Phosphorimager analysis for visualization of the bound radiolabeled protein (upper two rows). Graph of the results of multiple independent repetitions of the binding assay shown in A and B, with duplicate points in each repetition. Standard error bars are shown (n = 6 for WT, 3 for DSM). D) Wild-type (WT) and D-site-mutant (DSM) versions of SMTNL2 were tested as substrates for in vitro phosphorylation by active MAPKs. E) As shown in D, 0.25 μM of each GST-SMTNL2 protein was incubated with enzyme (0, 0.5 or 5 milliunits JNK1-3 or 0, 1, or 10 units ERK2) and [γ-32]ATP for 20 min. Samples were resolved on a 10% SDS-polyacrylamide gel and visualized by autoradiography.
