Figure 6.
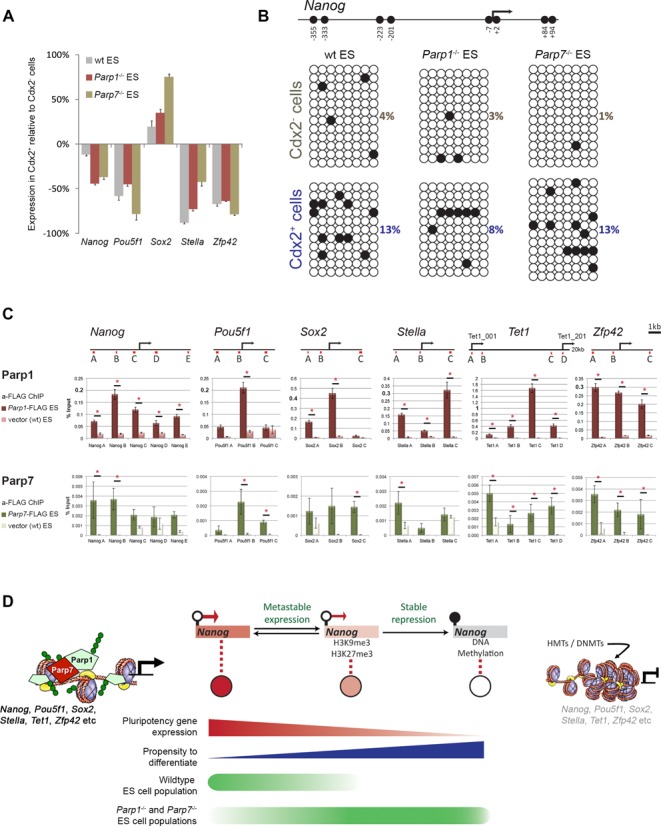
Parp1 and Parp7 contribute to maintaining the metastable state of pluripotency. (A) RT-qPCR analysis of pluripotency gene expression in wildtype (wt), Parp1-/- and Parp7-/- ES cells cultured in ES cell conditions and FACS-sorted into Cdx2-negative (Cdx2−) and -positive (Cdx2+) populations. Expression changes are calculated as the percentage difference between Cdx2+ and Cdx2− cell populations. (B) Bisulphite sequencing profiles of the Nanog locus (as depicted in Figure 5B) in the Cdx2− and Cdx2+ fractions of wt, Parp1-/- and Parp7-/- ES cells. (C) ChIP analysis of pluripotency locus occupancy by Parp1 and Parp7 using ES cells stably expressing C-terminally FLAG-tagged Parp1 and Parp7 constructs at approximately equal levels to endogenous proteins (Supplementary Figure S5). Chromatin was cross-linked with formaldehyde (for Parp1) and additionally with 2 mM di(N-succinimidyl)glutarate (for Parp7). For Parp1, ChIP was also performed against the endogenous protein on wt ES cells (Supplementary Figure S6). As controls, ChIPs against FLAG on wt ES cells, and with isotype-matched IgG on Parp1-FLAG/Parp7-FLAG cell lines were performed (Supplementary Figure S6). Schematic diagrams show the genomic locations of primer sets (red lines) used. All primers are given in the Supplementary Material. (*P < 0.05) (D) Model of Parp1 and Parp7 function in maintaining ES cell pluripotency. ES cells are normally in a dynamic equilibrium between higher and lower states of potency that are characterized by distinct histone modifications at key loci such as Nanog, Stella, Pecam1 and Zfp42. Parp1 and Parp7 preserve this dynamic equilibrium; in their absence, ES cells are more likely to acquire epigenetic repressive marks at pluripotency gene loci, including DNA methylation, which commits them towards differentiation. Green circles depict ADP-ribose moieties, which may be found on Parp1 and Parp7 themselves (auto-PARylation) or deposited on the immediate histone environment. Since PAR moieties introduce negative charges, a scenario of gentle electrostatic repulsion to relax the DNA fibre can be imagined as a mechanism to preserve an open chromatin structure or to repel binding of repressive factors. HMTs = histone methyltransferases, DNMTs = DNA methyltransferases.
