Figure 2.
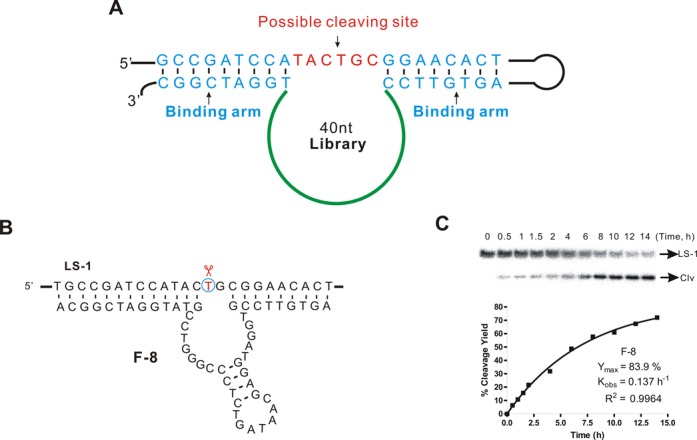
In vitro selection of DNA-cleaving DNAzymes. (A) Structure of the randomized library used for in vitro selection. A 40-nt randomized region was flanked by two binding arms that hybridized perfectly to the substrate strand containing the single-stranded target sequence TACTGC. (B) Structure of the in vitro-selected DNAzyme F-8. The thymidine at the cleavage site is circled. (C) PAGE analysis of an F-8 trans cleavage assay under single-turnover conditions (about 1 μM deoxyribozyme combined with 10 nM substrate) at 37°C in 50 mM HEPES (pH 7.4) containing 400 mM NaCl, 100 mM KCl, 10 mM MgCl2, 7.5 mM MnCl2 and 50 μM CuCl2. Representative results are shown for a 14-h time course; the observed rate constant (kobs ∼ 0.14 h−1, R2 = 0.9964) and maximum cleavage yield (Ymax) were derived using non-linear regression.
