Figure 6.
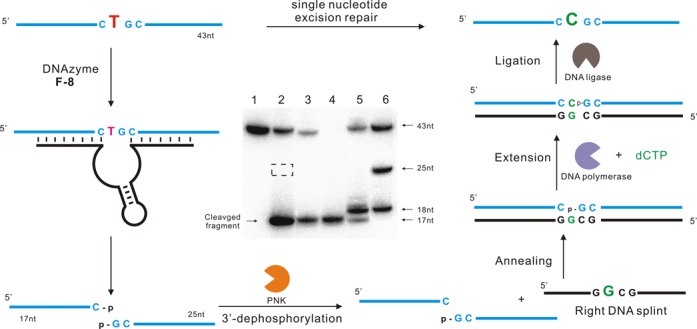
Conversion of a single deoxybonucleotide T (red, left) in a DNA strand to a deoxyribonucleotide C (green, right) using a combination of DNAzyme F-8 and natural enzymes. DNA substrate was cleaved by F-8 to generate both 3′- and 5′-phosphate termini. Then the 3′-phosphate group was removed by T4 polynucleotide kinase (PNK), and a right DNA splint was introduced. Subsequent extension and ligation restored the original DNA except for a substitution of T with C. Inset: PAGE analysis of the various stages of the single-nucleotide excision repair process. Lane 1, the original 43-nt 5′-32P-labeled DNA substrate. Lane 2, the upstream cleavage product generated by F-8. The cleaved fragment migrates slightly faster than the corresponding 18-nt marker because of its 3′-phosphate group. The dotted box indicates the expected position of the downstream cleavage product, which was invisible on the autoradiogram since it contained no 32P. Lane 3, the cleavage product after dephosphorylation with PNK. Lane 4, 5′-32P-labeled marker that comigrates with the 17-nt upstream cleavage fragment. Lane 5, the reaction products obtained after purifying the cleavage fragments, annealing them with right DNA splint, extending them by a C (18 nt) using DNA polymerase, and finally ligating to generate full-length substrate (43 nt). Lane 6, synthetic DNA markers. Cleavage assays were under standard single-turnover conditions (about 1 μM deoxyribozyme combined with 10 nM substrate).
