Figure 1.
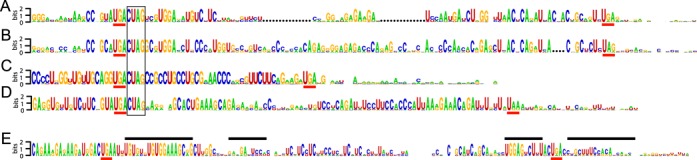
WebLogo representation showing the nucleotide conservation of the five readthrough candidates examined. Representation: (A) for OPRK1, (B) for OPRL1, (C) for MAPK10, (D) for AQP4 and (E) for SACM1L. Dotted areas in A and B represent gaps in the alignment between OPRK1 and OPRL1. The stop codons of the main ORFs and the readthrough extensions are underlined in red. The regions in SACM1L mRNA proposed to be involved in a stem-loop secondary structure are indicated by black bars above the conservation plot in E. The highly conserved tetranucleotide motif CUAG following the main ORF stop codon in OPRK1, OPRL1, AQP4 and MAPK10 is boxed.
