Figure 4.
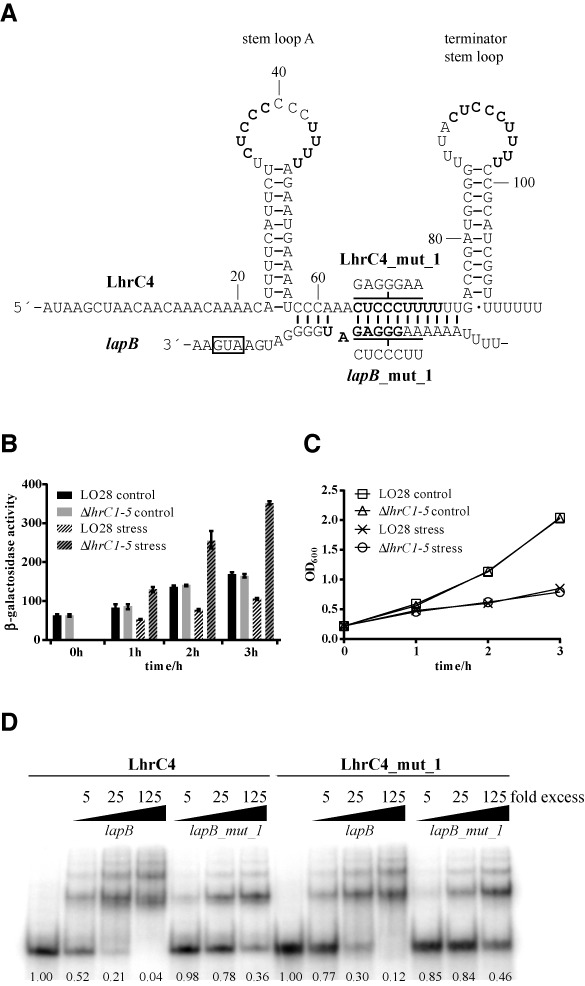
Predicted LhrC and lapB mRNA interaction. (A) According to the RNApredator software the single-stranded stretch between the two stem loops of LhrC binds to lapB mRNA thereby blocking the Shine–Dalgarno sequence (shown in bold). LhrC4 is shown as a representative of the five LhrC copies. Three similar sequences CUCCC(…)UUUU in loop A, the single-stranded stretch and the terminator loop, repectively, are marked in bold. The sequences mutated in LhrC4_mut_1 and lapB_mut_1 are shown. Lines above and below LhrC4 and lapB, respectively, indicate the regions mutated in the mut_1 variants. (B) LhrC-mediated down-regulation of lapB-lacZ expression. β-gal activities were assessed in wild type and ΔlhrC1–5 strains carrying pC-lapB-lacZ. Under non-stress conditions β-gal activity of wild type and mutant was comparable at all tested time points. After cefuroxime stress, β-gal activity in the wild type strain decreased whereas in ΔlhrC1–5 an increase was observed. After 2 and 3 h of stress, β-gal activity in the mutant was more than three times as high as in the wild type, pointing to a released negative regulation in ΔlhrC1–5. Results of the β-gal assay are the average of four biological replicates each conducted in technical duplicates. (C) Growth of ΔlhrC1–5 and wild type strains, tested in the β-gal assay shown in Figure 4(B), was comparable at all tested time points. (D) Gel mobility shift assay of the interaction between lapB mRNA and LhrC4. Labeled LhrC4 was shifted with increasing concentrations of lapB mRNA. Mutation of 7 nt in the lapB sequence predicted to be involved in the interaction (lapB_mut_1) reduced the interaction. Correspondingly mutated LhrC4 (LhrC4_mut_1) was still capable of lapB mRNA binding and could not compensate for the mutation in lapB_mut_1. The fraction of unbound LhrC is shown below each lane.
