Figure 1.
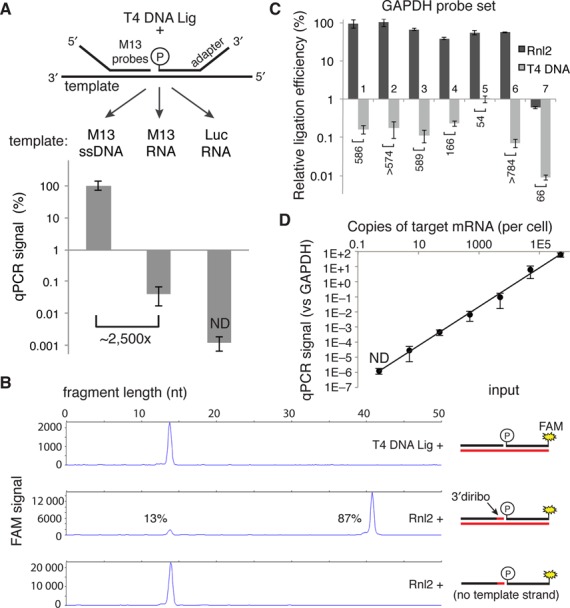
Measurement of T4 DNA Ligase and Rnl2 probe joining activity on DNA and RNA templates. (A) 500 nM of each adapter-M13 probe was annealed on the indicated 100 nM template (60°C for 10 min, 45°C for 30 min) in 1× T4 DNA ligase buffer. Complexes were then diluted 1000-fold into a 20 μl ligation reaction containing 5 U of T4 DNA ligase and incubated at 37°C for 30 min. Ligation products were then diluted 1000-fold into SYBR green master mix before undergoing PCR. qPCR signals are expressed as percent of the ssDNA template condition. (B) Capillary electrophoresis of ligation products. A FAM-labeled donor probe at 1 μM and the indicated hybrid diribo- or deoxyribo-acceptor oligo (10 μM) were annealed on a synthetic GAPDH RNA template (10 μM) or no template and then diluted 10-fold into the indicated ligation reaction and incubated for 30 min at 37°C. The product of this ligation was then diluted 10-fold in stopping buffer and measured by capillary electrophoresis. (C) Seven GAPDH probe sets were tested with the indicated ligase in the RASL assay, using 50 ng of prostate RNA as template in a 20 μl reaction. After 10 cycles of pre-amplification, the product of each probe set was separately analyzed by qPCR using probe set-specific nested detection primers. (D) Sensitivity of an Rnl2-based RASL assay, determined by serial dilution of a synthetic M13 RNA template into a background of 50 ng prostate RNA (the equivalent of ∼2000 cells). Data reported are the mean of at least three independent replicates; error bars are S.E.M. ND, not detected.
