Figure 2.
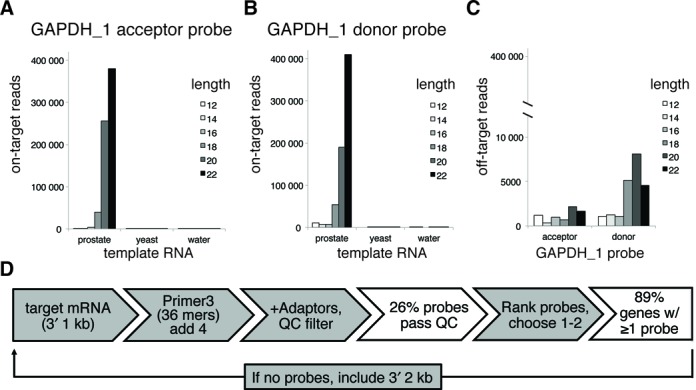
Probe length analysis and design pipeline. (A) and (B) Independent probe sets targeting GAPDH and M13 were synthesized as both donor and acceptor oligos ranging in target sequence length from 12 to 22 nucleotides; the junction between acceptor and donor probes was kept constant. These oligos were pooled such that the total concentration of each pooled probe set was 5 nM in the final RASL-seq assay. The lengths of correctly (on target) ligated donor or acceptor probes were determined by deep sequencing, and each length's contribution to the total number of observed read counts was tabulated. Data from the GAPDH_1 probe sets are shown when prostate RNA, yeast RNA or no template was included in the assay. (C) The contribution of each probe length to off-target ligations (defined as inappropriate ligation between different probe sets) is shown for the prostate RNA template condition. The scale is different because the off-target ligations are relatively rare compared to the on-target ligations. (D) The probe design pipeline takes the 1 kb 3′-sequence upstream of the poly(A) and uses Primer3 to generate candidate 36 nt RASL probe sequences (reverse complement of mRNA sense strand). After extension of 4 nt in the poly(A) direction, splitting of the 40mer into acceptor and donor, and appending of appropriate adapters, the properties of the adaptor-appended probe oligos are calculated. Probes are then filtered through a quality control (QC) step (‘Materials and Methods’ section), which removes ∼75% of the candidates (based on analysis of 1000 transcripts). Successfully filtered probes are finally ranked by a combination of proximity to poly(A) and Primer3 penalty (based on the original 36 nt candidate probe), such that the best probe sets can be selected for production. Gray boxes denote processes and white boxes denote the outcomes.
