Figure 4.
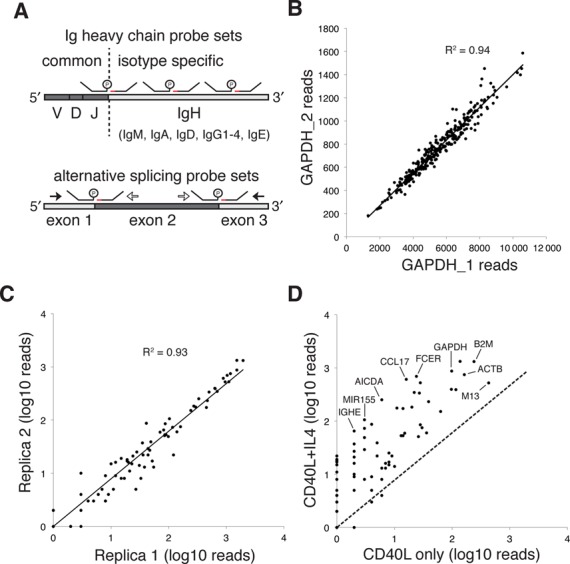
Design of B cell probes and assay validation. (A) At least two probe sets were designed to target the constant region of each antibody isotype heavy chain. In addition, a donor probe targeting the J-segment of the variable domain was used as a common donor, capable of ligation with any isotype-specific acceptor probe. For transcripts with important splice variants, RASL probes were designed so that ligation junctions aligned with exon boundaries. Probes outside an alternatively included exon are referred to as ‘common’ (filled black arrows) and internal are referred to as ‘long’ (open arrows). Common probes will ligate with each other when the exon is excluded and with long probes when the exon is included. (B) RASL-seq data for GAPDH_1 and GAPDH_2 probe sets across one 384 well plate. (C) RASL-seq read counts from all B cell probes corresponding to two replicate negative control wells (no drug treatment), present on one 384 well plate. (D) RASL-seq read counts from all B cell probes corresponding to two negative control wells, one of which did not include IL-4 treatment. IGHE, immunoglobulin heavy constant epsilon; MIR155, miR-155 host gene; AICDA, activation-induced cytidine deaminase; CCL17, chemokine (C-C motif) ligand 17; FCER, high affinity receptor for the Fc fragment of IgE; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; M13, synthetic spike in transcript; B2M, beta-2-microglobulin; ACTB, beta actin. A pseudocount of 1 was added to the read counts of (C) and (D) so that all data points could be shown on the log–log plot.
