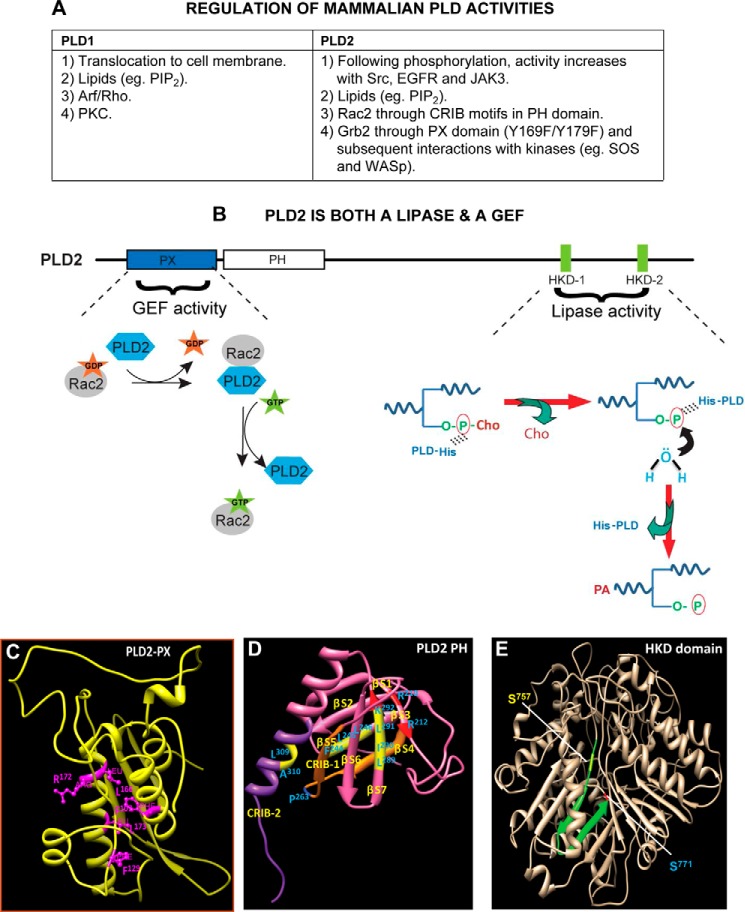
FIGURE 2.
Regulation of PLD enzymatic activities. A, list of specific regulation(s) of PLD1 and PLD2, the most studied mammalian isoforms. PIP2, phosphatidylinositol 4,5-bisphosphate; CRIB motif, Cdc42/Rac interactive binding motif. B, phospholipase D2 is a dual enzyme that catalyzes a lipase activity, as well as a guanine nucleotide exchange. Shown are the N-terminal PLD2-PX (where part of the GEF activity resides) and the C-terminal HKD1/2 domains (where lipase activity resides). For the GEF reaction, PLD2 causes Rac2-based GDP dissociation upon interaction with Rac2-GDP. In a second step, PLD2 stabilizes nucleotide-free Rac2 until GTP binds, after which PLD2 is released from the complex, leading to the activation of Rac2. For the lipase reaction, the catalytic HKD motifs of PLD2 fold around the substrate phosphatidylcholine (P-Cho). In the first step, a phosphatidyl-histidine intermediate is generated due to a nucleophilic attack of the histidine of the lipases on the phosphate of phosphatidylcholine. In the next step, the hydroxyl group of water attacks the phosphatidyl-histidine intermediate, leading to the formation of phosphatidic acid, at which time the enzyme is regenerated for the next cycle of PC breakdown. C–E, schematic drawings of main domains in the PLD2 structure. C, ribbon model of PLD2-PX domain noting key amino acids needed for the GEF activity. D, ribbon model of PLD2-PH domain that includes CRIB-1 and CRIB-2 needed for interaction with small GTPases (e.g. Rac2). E, ribbon model of PLD2-HKD domain. Serine residues that are mutated for inhibitor studies but that retain lipase activity are highlighted. The structures in C–E were generated by using protein prediction servers such as I-TASSER and Phyre (52). Once the structures were obtained, they were validated using biochemistry data available from both my laboratory and those of published authors in the field.