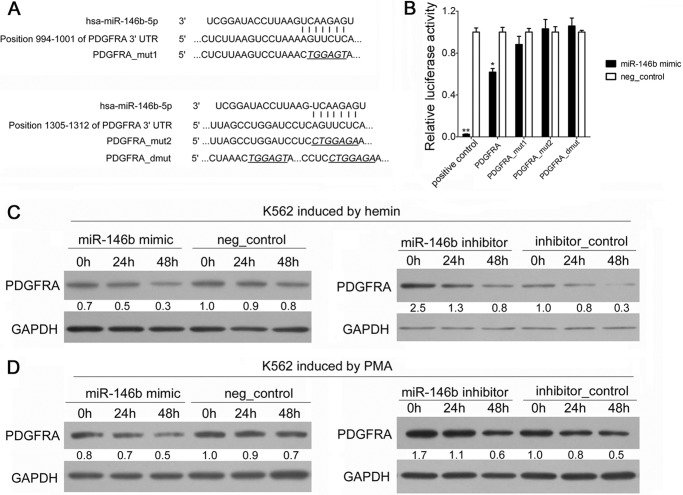
FIGURE 3.
PDGFRA mRNA was identified and validated as a direct target of miR-146b. A, computer prediction of conserved binding sites within the 3′UTR of PDGFRA mRNA for miR-146b. The mutated base sequences in the luciferase reporter assays are italicized and underlined. B, relative luciferase activity of the indicated PDGFRA reporter constructs. Error bars represent the standard deviation obtained from three independent experiments. *, p < 0.05; **, p < 0.01. C, immunoblot analysis of PDGFRA in the K562 cells transfected with scramble or miR-146b mimics or inhibitors, followed by hemin induction for 0, 24, or 48 h. D, immunoblot analysis of PDGFRA in K562 cells transfected with scrambled or miR-146b mimics or inhibitors, and PMA induction for 0, 24, or 48 h. For all Western blots, GAPDH antibody was used to assess equal protein loading. The signal in each lane was quantified using Gelpro32 software, and the ratio of PDGFRA to GAPDH was determined. PDGFRA expression was calculated as the relative fold with respect to its expression in the negative (neg)-control-transfected cells before differentiation induction.