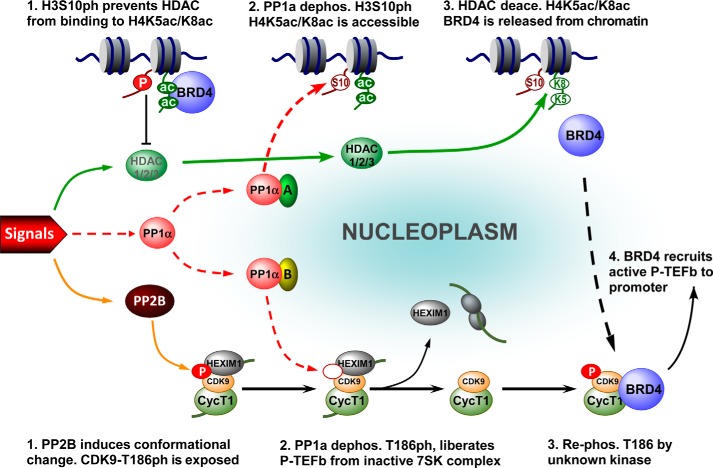
FIGURE 8.
A model depicting the role of the PP1α signaling pathway in coordinating the activation of BRD4 and P-TEFb for inducible gene expression. Top: (1) in the absence of external signals, almost all BRD4 is locked up in nucleosomes containing histone modifications of H4K5ac/K8ac and H3S10ph. H3S10ph prevents HDAC from deacetylating H4K5ac/K8ac, thereby prohibiting the inappropriate activation of BRD4 (2). Upon stimulation, the external signals (left) trigger the activation of PP1α, which in association with partner A, dephosphorylates H3S10ph (3). Subsequently, the signal-activated HDAC1/2/3 deacetylate H4K5ac/K8ac, which leads to BRD4 release from chromatin. Bottom: (1) meanwhile, the signal-activated phosphatase PP2B induces a conformational change in the inactive 7SK complex to expose CDK9 T186ph (2). Consequently, PP1α with partner B is able to dephosphorylate CDK9 at Thr186, thereby liberating P-TEFb from the inactive 7SK complex (14) (3). Through selective association with the transcriptionally active form of P-TEFb that has its CDK9 T-loop re-phosphorylated by unknown kinase(s), (4) the released BRD4 mediates the recruitment of P-TEFb to the promoter-proximal region to enhance the processivity of RNA Pol II for productive elongation.