FIGURE 1.
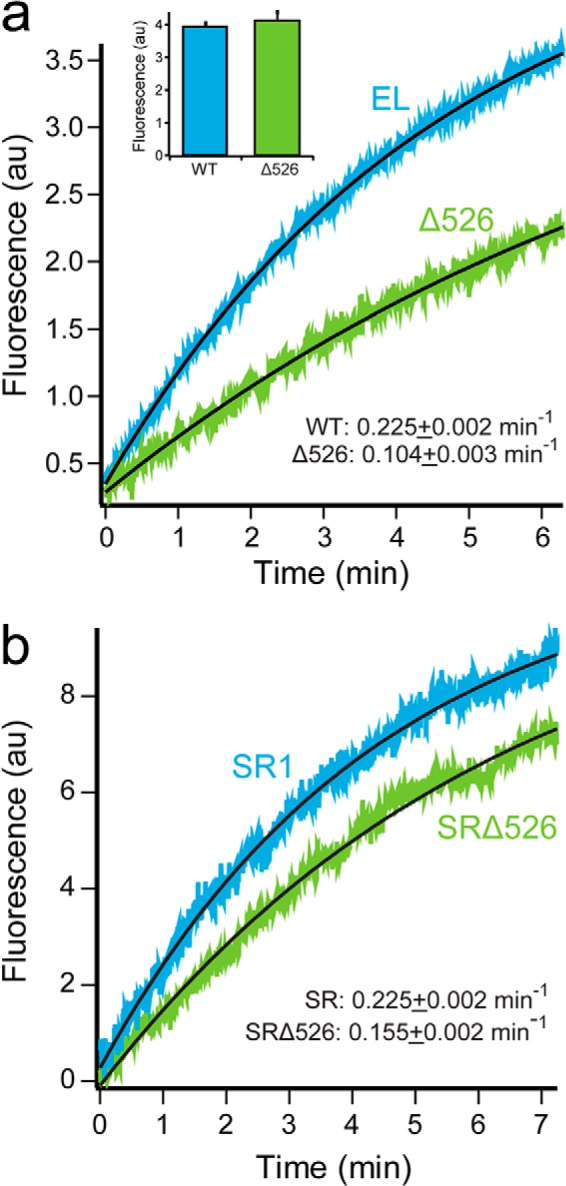
Presence of the GroEL C-terminal tails enhances protein folding. a, folding of Rubisco by cycling GroEL-GroES was monitored by an increase in tryptophan fluorescence. Chemically denatured, wild-type Rubisco (100 nm) was bound to GroEL (200 nm), wild-type (EL; blue), or C-terminal deletion (Δ526; green) and rapidly mixed with an equal volume of excess GroES (400 nm) and ATP (2 mm) in a stopped-flow apparatus. Curves were fit to a single-exponential rate law (black line), resulting in observed rate constants of 0.225 ± 0.002 min−1 for GroEL and 0.104 ± 0.003 min−1 for Δ526. In each case, the overall folding yield was examined by allowing each folding reaction to run to completion (∼30 min), followed by a measurement of the total tryptophan fluorescence (inset). b, folding of Rubisco inside stable single ring-GroES complexes monitored by an increase in tryptophan fluorescence. Chemically denatured wild-type Rubisco (100 nm) was bound to full-length SR1 (SR, blue) or Δ526 (SRΔ526, green) single-ring variants of GroEL (300 nm) and rapidly mixed with an equal volume of excess GroES (600 nm) and ATP (2 mm) in a stopped-flow apparatus. Curves were fit to single-exponential rate laws, yielding observed rate constants of 0.225 ± 0.002 min−1 for SR1 and 0.155 ± 0.002 min−1 for SRΔ526. n = 10 replicates, with a 100-ms sampling time. AU, arbitrary units.
